Decoding Hepatocellular Carcinoma: A Comprehensive Guide to Non-Coding RNA Sequencing Analysis
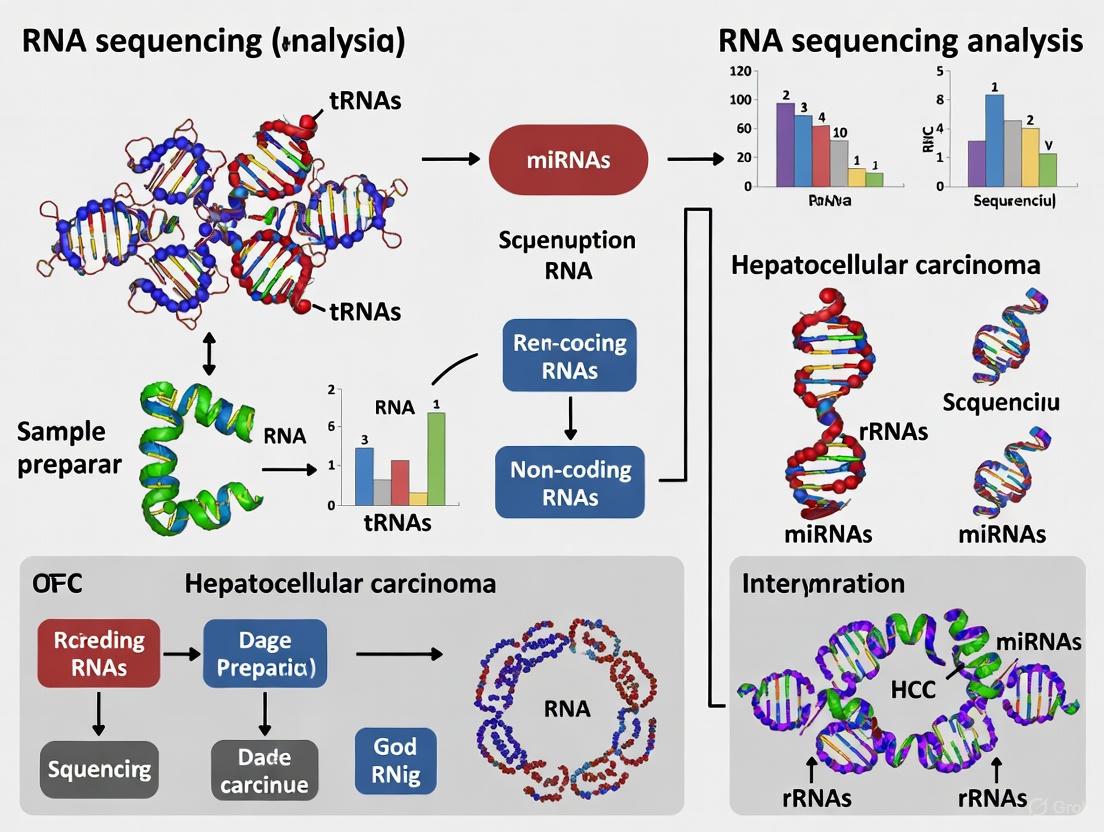
This article provides a comprehensive exploration of RNA sequencing for profiling non-coding RNAs (ncRNAs) in hepatocellular carcinoma (HCC) tissues.
Decoding Hepatocellular Carcinoma: A Comprehensive Guide to Non-Coding RNA Sequencing Analysis
Abstract
This article provides a comprehensive exploration of RNA sequencing for profiling non-coding RNAs (ncRNAs) in hepatocellular carcinoma (HCC) tissues. It covers the foundational biology of key ncRNAs—including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs)—and their roles in HCC progression, drug resistance, and the tumor microenvironment. The review details state-of-the-art methodological approaches, such as single-cell and bulk RNA-seq integration and machine learning for biomarker discovery. It further addresses common analytical challenges and offers optimization strategies, concluding with a critical evaluation of biomarker validation techniques and the translational potential of ncRNA signatures for diagnosis, prognosis, and novel therapeutics, aiming to bridge the gap between computational analysis and clinical application for researchers and drug development professionals.
The Landscape of Non-Coding RNAs in HCC Pathogenesis
Hepatocellular carcinoma (HCC) represents a significant global health challenge, ranking as the sixth most common malignancy and the fourth leading cause of cancer-related mortality worldwide [1]. The molecular pathogenesis of HCC involves the accumulation of genetic and epigenetic alterations that drive the transformation of hepatocytes, with non-coding RNAs (ncRNAs) emerging as crucial regulators in this process [2] [3]. These RNA transcripts, which lack protein-coding capacity, constitute the majority of the human transcriptome and play essential roles in regulating gene expression at multiple levels [4]. In the context of a broader thesis on RNA sequencing analysis of non-coding RNAs in HCC tissues, this application note provides a comprehensive overview of the classification, characteristics, and experimental approaches for studying three major ncRNA categories: microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). Understanding the distinct properties and functions of these ncRNA classes is fundamental to elucidating their roles in HCC pathogenesis and identifying novel diagnostic biomarkers and therapeutic targets.
Classification and Characteristics of HCC-Associated ncRNAs
Non-coding RNAs are broadly categorized based on molecular size and structural characteristics. The three principal classes implicated in HCC pathogenesis—miRNAs, lncRNAs, and circRNAs—exhibit distinct biogenesis pathways, structural features, and functional mechanisms [3] [5] [6].
MicroRNAs (miRNAs)
Characteristics and Biogenesis: miRNAs are small endogenous non-coding RNAs approximately 19-30 nucleotides in length that function as post-transcriptional regulators of gene expression [1]. The biogenesis of miRNAs begins with RNA polymerase II-mediated transcription of primary miRNA (pri-miRNA) transcripts. These pri-miRNAs are processed in the nucleus by the Drosha-DGCR8 complex to form precursor miRNAs (pre-miRNAs) of approximately 70-100 nucleotides with hairpin structures. After export to the cytoplasm via exportin-5, pre-miRNAs are cleaved by Dicer enzyme to generate mature miRNA duplexes. One strand of this duplex is loaded into the RNA-induced silencing complex (RISC), where it guides target recognition through complementary base pairing with messenger RNAs (mRNAs), leading to translational repression or mRNA degradation [1].
Functional Mechanisms in HCC: In HCC, miRNAs function as critical regulators of oncogenic and tumor-suppressive pathways. They are conventionally classified as oncomiRs (oncogenic miRNAs) or tumor-suppressor miRNAs (TS-miRs) based on their target genes and biological effects [2] [1]. For instance, miR-221, one of the most investigated oncomiRs in HCC, promotes tumor growth by targeting the DDIT4/mTOR pathway and interfering with apoptosis through regulation of PTEN and TIMP3 via AKT pathway activation [2]. Conversely, tumor-suppressor miRNAs such as miR-122, miR-29, and miR-195 are frequently downregulated in HCC. miR-122 attenuates HCC progression, and its delivery in animal models suppresses liver tumor development [2]. miR-29 targets multiple oncogenic pathways including IGF2BP1, VEGFA, and BCL2, while miR-195 impedes angiogenesis by targeting VEGF, VAV2, and CDC42 [2].
Table 1: Key miRNAs Dysregulated in Hepatocellular Carcinoma
| miRNA | Expression in HCC | Category | Validated Targets | Functional Effects |
|---|---|---|---|---|
| miR-221 | Upregulated | OncomiR | DDIT4, PTEN, TIMP3 | Promotes proliferation, inhibits apoptosis [2] |
| miR-122 | Downregulated | TS-miR | Multiple | Suppresses tumor development [2] |
| miR-29 | Downregulated | TS-miR | IGF2BP1, VEGFA, BCL2 | Contrasts proliferation, angiogenesis [2] |
| miR-195 | Downregulated | TS-miR | VEGF, VAV2, CDC42 | Impedes angiogenesis [2] |
| miR-101 | Downregulated | TS-miR | ROCK | Inhibits metastasis [2] |
| miR-497 | Downregulated | TS-miR | Rictor/AKT pathway | Contrasts proliferation, invasion, metastasis [2] |
Long Non-Coding RNAs (lncRNAs)
Characteristics and Classification: Long non-coding RNAs (lncRNAs) are defined as RNA transcripts exceeding 200 nucleotides in length that lack protein-coding potential [3] [7]. These molecules exhibit tissue-specific expression patterns and are classified based on their genomic location relative to protein-coding genes: (1) sense lncRNAs, which overlap with exons of protein-coding genes; (2) antisense lncRNAs, transcribed from the opposite strand of protein-coding genes; (3) bidirectional lncRNAs, positioned head-to-head with protein-coding genes; (4) intronic lncRNAs, derived entirely from introns; and (5) intergenic lncRNAs, located between protein-coding genes [3].
Functional Mechanisms: LncRNAs exert their biological functions through diverse molecular mechanisms, including chromatin modification, transcriptional regulation, and post-transcriptional processing [3] [7]. They can function as signals, decoys, guides, or scaffolds in regulating gene expression. In HCC, numerous lncRNAs demonstrate aberrant expression and contribute to tumorigenesis through various pathways. For example, lncRNA HULC is upregulated in HCC and promotes tumor growth, metastasis, and drug resistance [7]. LncRNA H19, one of the first identified lncRNAs, restricts organ growth by decreasing IGF2 expression [7]. The lncRNA UCA1 promotes cell proliferation, while GAS5 inhibits cancer cell proliferation and activates apoptosis through CHOP and caspase-9 signaling pathways [8].
Table 2: Key Long Non-Coding RNAs in Hepatocellular Carcinoma
| LncRNA | Expression in HCC | Functional Role | Molecular Mechanisms | Clinical Relevance |
|---|---|---|---|---|
| HULC | Upregulated | Oncogenic | Promotes growth, metastasis, drug resistance [7] | Potential therapeutic target |
| H19 | Upregulated | Oncogenic | Decreases IGF2 expression [7] | Early discovered lncRNA |
| UCA1 | Upregulated | Oncogenic | Promotes proliferation [8] | Diagnostic biomarker potential |
| GAS5 | Downregulated | Tumor suppressor | Activates CHOP, caspase-9 pathways [8] | Promotes apoptosis |
| MALAT1 | Upregulated | Oncogenic | Promotes aggressive tumor phenotypes [8] | Associated with progression |
| LINC00152 | Upregulated | Oncogenic | Regulates CCDN1 [8] | Diagnostic biomarker |
Circular RNAs (circRNAs)
Characteristics and Biogenesis: Circular RNAs are a novel class of endogenous non-coding RNAs characterized by covalently closed continuous loop structures formed through back-splicing events, which confer exceptional stability due to resistance to exonuclease-mediated degradation [5] [6]. These molecules lack 5' caps and 3' polyadenylated tails and are classified into three main categories based on their genomic origin: exonic circRNAs (EcircRNAs), which consist primarily of exonic sequences; circular intronic RNAs (ciRNAs), derived from intronic sequences; and exon-intron circRNAs (EIciRNAs), which contain both exonic and intronic regions [5].
Functional Mechanisms: CircRNAs perform diverse biological functions, with the most well-characterized being their role as competitive endogenous RNAs (ceRNAs) that function as miRNA "sponges," sequestering miRNAs and preventing them from binding to their target mRNAs [5] [6]. Additional functions include regulation of transcription and alternative splicing, interaction with RNA-binding proteins (RBPs), and serving as templates for translation. In HCC, numerous circRNAs exhibit dysregulated expression and contribute to tumor progression. For example, circ_0008450 promotes proliferation, invasion, and migration while inhibiting apoptosis via regulation of miR-548p [5]. Conversely, circADAMTS14 inhibits HCC progression by regulating the miR-572/RCAN1 axis [5]. CDR1as, one of the most extensively studied circRNAs, contains multiple binding sites for miR-7 and functions as a molecular sponge, influencing HCC development [5].
Table 3: Key Circular RNAs in Hepatocellular Carcinoma
| circRNA | Expression in HCC | Functional Role | Molecular Mechanisms | Regulatory Axis |
|---|---|---|---|---|
| circ_0008450 | Upregulated | Oncogenic | Promotes proliferation, invasion, migration; inhibits apoptosis [5] | miR-548p |
| circRNA-104718 | Upregulated | Oncogenic | Promotes proliferation, invasion; inhibits apoptosis [5] | miRNA-218-5p/TXNDC5 |
| circADAMTS14 | Downregulated | Tumor suppressor | Inhibits proliferation, invasion, migration; promotes apoptosis [5] | miR-572/RCAN1 |
| circRNA-5692 | Downregulated | Tumor suppressor | Inhibits proliferation, invasion, migration [5] | miR-328-5p/DAB2IP |
| CDR1as | Upregulated | Oncogenic | Functions as molecular sponge for miR-7 [5] | miR-7 |
| cSMARCA5 | Downregulated | Tumor suppressor | Functions as molecular sponge [5] | Multiple miRNAs |
Experimental Protocols for ncRNA Analysis in HCC Research
RNA Sequencing for ncRNA Profiling
Objective: To comprehensively identify and quantify miRNAs, lncRNAs, and circRNAs in HCC tissues and matched non-tumor liver tissues.
Workflow:
- RNA Extraction: Isolate total RNA from fresh-frozen or RNAlater-preserved HCC tissues and matched non-tumor liver tissues using miRNeasy Mini Kit (QIAGEN), which efficiently recovers both small and large RNA species [9] [8].
- RNA Quality Control: Assess RNA integrity using Agilent Bioanalyzer RNA Nano Chip, ensuring RIN (RNA Integrity Number) >7.0 for sequencing applications.
- Library Preparation:
- For miRNA sequencing: Use 1μg total RNA with QIAseq miRNA Library Prep Kit (QIAGEN) featuring unique molecular identifiers (UMIs) to eliminate amplification bias [9].
- For lncRNA/circRNA sequencing: Deplete ribosomal RNA using NEBNext rRNA Depletion Kit, followed by library preparation with NEBNext Ultra II Directional RNA Library Prep Kit for Illumina.
- For circRNA-specific analysis: Treat RNA with RNase R to degrade linear RNAs and enrich for circular transcripts [6].
- Sequencing: Perform high-throughput sequencing on Illumina platforms (e.g., NovaSeq 6000) with recommended depths: 50 million single-end 75bp reads for miRNA; 100 million paired-end 150bp reads for lncRNA/circRNA [6].
- Bioinformatic Analysis:
- miRNA analysis: Process raw data with Cutadapt to remove adapters, then align to miRBase using Bowtie2. Quantify expression with featureCounts and perform differential expression analysis using DESeq2.
- lncRNA analysis: Align reads to human genome (GRCh38) using STAR aligner. Assemble transcripts with StringTie and identify novel lncRNAs using CPC2 and FEELnc.
- circRNA analysis: Detect circRNAs using CIRI2 and CIRCexplorer2 algorithms, requiring at least two unique back-spliced reads for circRNA identification [6].
Figure 1: RNA Sequencing Workflow for ncRNA Profiling in HCC Tissues
Validation by Quantitative Real-Time PCR (qRT-PCR)
Objective: To validate sequencing results through targeted quantification of differentially expressed ncRNAs.
Protocol:
- cDNA Synthesis:
- For miRNA: Use miscript II RT Kit (QIAGEN) with poly(A) tailing and universal reverse transcription primer.
- For lncRNA/circRNA: Use RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) with random hexamers and oligo(dT) primers [8].
- qRT-PCR Reaction:
- Prepare reactions with PowerTrack SYBR Green Master Mix (Applied Biosystems)
- Use gene-specific primers with the following design considerations:
- miRNA: Stem-loop primers for mature miRNA quantification
- circRNA: Divergent primers spanning back-splice junctions to specifically amplify circular isoforms
- Perform amplification in triplicate on ViiA 7 Real-Time PCR System (Applied Biosystems) with the following cycling conditions: 95°C for 10min, followed by 40 cycles of 95°C for 15sec and 60°C for 1min [8].
- Data Analysis: Calculate relative expression using the 2^(-ΔΔCt) method with appropriate normalization:
- miRNA: U6 snRNA or RNU44
- lncRNA/circRNA: GAPDH or β-actin [8]
Functional Characterization of ncRNAs
Objective: To investigate the biological roles of specific ncRNAs in HCC pathogenesis.
Gain-of-Function and Loss-of-Function Studies:
- Expression Modulation:
- Overexpression: Clone full-length ncRNA sequences into pcDNA3.1 or lentiviral vectors (e.g., pLenti-CMV-GFP-Puro)
- Knockdown: Design antisense oligonucleotides (ASOs) for lncRNAs or miRNA inhibitors (antagomiRs) using locked nucleic acid (LNA) technology
- In Vitro Functional Assays:
- Proliferation: MTT assay, colony formation assay
- Apoptosis: Annexin V/PI staining with flow cytometry
- Migration/Invasion: Transwell assays with Matrigel coating
- Mechanistic Studies:
- miRNA Sponging: Dual-luciferase reporter assays with wild-type and mutant target sequences
- Protein Interactions: RNA immunoprecipitation (RIP) using Magna RIP Kit (Millipore)
- Subcellular Localization: RNA fluorescence in situ hybridization (RNA-FISH)
The Scientist's Toolkit: Essential Research Reagents
Table 4: Essential Research Reagents for HCC ncRNA Studies
| Category | Product/Kit | Manufacturer | Application | Key Features |
|---|---|---|---|---|
| RNA Extraction | miRNeasy Mini Kit | QIAGEN | Simultaneous purification of miRNA and total RNA | Maintains miRNA integrity, high purity [9] [8] |
| cDNA Synthesis | RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific | cDNA synthesis for lncRNA/circRNA | High efficiency, includes RNase inhibitor [8] |
| qRT-PCR | PowerTrack SYBR Green Master Mix | Applied Biosystems | Quantitative PCR detection | Optimized for difficult templates, low background [8] |
| Library Prep | QIAseq miRNA Library Kit | QIAGEN | miRNA sequencing library | Unique Molecular Identifiers (UMIs) [9] |
| Ribodepletion | NEBNext rRNA Depletion Kit | New England Biolabs | rRNA removal for RNA-seq | Efficient ribosomal RNA removal |
| circRNA Enrichment | RNase R | Epicentre | circRNA enrichment | Degrades linear RNAs, enriches circular forms [6] |
| Functional Studies | Locked Nucleic Acids (LNA) | Qiagen/Exiqon | miRNA inhibition | Enhanced binding affinity, nuclease resistance |
| Urushiol II | Urushiol II|Catechol Derivative|For Research Use | Urushiol II is a natural catechol derivative for antimicrobial, anticancer, and materials science research. For Research Use Only. Not for human consumption. | Bench Chemicals | |
| Astragaloside II | Astragaloside II, CAS:84676-89-1, MF:C43H70O15, MW:827.0 g/mol | Chemical Reagent | Bench Chemicals |
ncRNA Regulatory Networks in HCC
The complex interplay between different ncRNA classes forms intricate regulatory networks that drive HCC pathogenesis. Understanding these networks is essential for comprehending the molecular basis of hepatocellular carcinoma and identifying therapeutic intervention points.
Figure 2: ncRNA Regulatory Network in Hepatocellular Carcinoma
This regulatory network illustrates the complex interactions between different ncRNA classes in HCC. circRNAs such as CDR1as function as miRNA sponges, sequestering miRNAs and preventing them from inhibiting their target tumor suppressor genes [5]. Similarly, lncRNAs like HULC can act as competitive endogenous RNAs, binding to miRNAs and modulating their availability [7]. Meanwhile, specific miRNAs directly target key oncogenes or tumor suppressors, creating a finely balanced regulatory system that becomes disrupted during hepatocarcinogenesis. Understanding these networks provides insights into potential therapeutic interventions that could restore normal regulatory balance in HCC cells.
The comprehensive characterization of ncRNAs in hepatocellular carcinoma represents a crucial frontier in cancer research with significant implications for diagnostic and therapeutic development. miRNAs, lncRNAs, and circRNAs each possess distinct characteristics and contribute to HCC pathogenesis through diverse yet interconnected molecular mechanisms. The experimental protocols outlined in this application note provide a framework for systematic investigation of these RNA molecules in HCC tissues, from initial discovery through functional validation. As research in this field advances, the integration of ncRNA profiling into clinical practice holds promise for improving early detection, prognostic stratification, and treatment selection for HCC patients. Furthermore, the unique properties of circRNAs, particularly their stability and tissue-specific expression patterns, position them as particularly promising candidates for future diagnostic and therapeutic applications. Continued investigation of ncRNA regulatory networks will undoubtedly yield novel insights into HCC biology and contribute to the development of more effective precision medicine approaches for this devastating malignancy.
Hepatocellular carcinoma (HCC) represents a significant global health challenge, characterized by high mortality rates and limited treatment options for advanced disease. The complexity of HCC is driven by substantial morphological, genetic, and epigenetic heterogeneity, which poses considerable challenges for developing effective targeted therapies [10]. In recent years, non-coding RNAs (ncRNAs) have emerged as crucial regulators of gene expression and cellular processes in carcinogenesis. Among these, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) have demonstrated significant roles in HCC pathogenesis, functioning as both oncogenic drivers and tumor suppressors [11].
Two ncRNAs of particular interest are miR-221 and the growth arrest-specific transcript 5 (GAS5). miR-221 is a well-characterized oncogenic miRNA frequently upregulated in HCC, where it promotes cell proliferation, migration, and invasion while inhibiting apoptosis [12] [13]. In contrast, GAS5 presents a more complex picture—traditionally considered a tumor suppressor in many cancers but demonstrating oncogenic functions in specific HCC contexts [14] [15]. This application note examines the dysregulated functions of these ncRNAs within the framework of RNA sequencing analysis of HCC tissues, providing experimental protocols and analytical frameworks for researchers investigating ncRNA roles in liver cancer.
Key ncRNAs in Hepatocellular Carcinoma
Oncogenic miR-221 in HCC
miR-221 represents one of the most consistently upregulated miRNAs in HCC, with demonstrated roles in multiple aspects of tumor progression. Clinical evidence shows miR-221 overexpression significantly correlates with advanced TNM stages, metastasis, and tumor capsular infiltration [12]. Functional studies confirm its role in enhancing cell growth, inhibiting apoptosis, and promoting invasive capabilities [12] [13].
Table 1: Oncogenic Functions of miR-221 in Hepatocellular Carcinoma
| Functional Role | Experimental Evidence | Target Genes/Pathways | Clinical Correlation |
|---|---|---|---|
| Cell Proliferation | Increased viability in Hep3B, HepG2, and SNU449 cells [12] | CDKN1B/p27, CDKN1C/p57 [12] | Tumor size, differentiation grade |
| Apoptosis Inhibition | Reduced caspase-3/7 activity; decreased apoptosis [12] | Unknown | Shorter time-to-recurrence |
| Migration & Invasion | Enhanced migratory/invasive abilities [13] | LIFR, MTSS1, FOXO3a [13] | Metastasis, capsular infiltration |
| Cell Cycle Progression | Increased S-phase population [12] | p27, p57 [12] | Advanced TNM stage [12] |
The Dual Nature of GAS5 in HCC
The lncRNA GAS5 presents a more complex picture in HCC, with evidence supporting both tumor-suppressive and oncogenic functions depending on context and molecular interactions. This apparent contradiction highlights the context-dependent nature of ncRNA functions in cancer biology.
Table 2: Dual Functions of GAS5 in Hepatocellular Carcinoma
| GAS5 Function | Expression Pattern | Molecular Mechanisms | Functional Outcomes |
|---|---|---|---|
| Tumor Suppressor | Downregulated in HCC tissues [16] | Sponging miR-182, upregulating ANGPTL1 [16] | Inhibits migration, invasion, and metastasis [16] |
| Oncogene | Upregulated in HCC, associated with poor survival [15] | Competing with miR-423-3p to regulate SMARCA4 [15] | Promotes tumor growth and proliferation [15] |
| Therapeutic Target | Modulated by UTMD-mediated transfection [16] | Acting as ceRNA for multiple miRNAs | Suppresses metastatic abilities [16] |
Experimental Protocols for ncRNA Functional Analysis
Protocol 1: Functional Validation of miR-221 Using Gain- and Loss-of-Function Studies
Objective: To determine the functional effects of miR-221 on HCC cell proliferation, apoptosis, and cell cycle progression.
Materials and Reagents:
- HCC cell lines (Hep3B, HepG2, SNU449)
- miR-221 mimic and inhibitor (e.g., from Dharmacon)
- Appropriate transfection reagent (e.g., Lipofectamine 3000)
- Cell viability assay kits (MTS tetrazolium or fluorimetric resorufin)
- Apoptosis detection kit (Annexin V/PI staining)
- Cell cycle analysis reagents (PI/RNase staining)
- Western blot equipment for target protein detection
Procedure:
- Cell Culture and Transfection:
- Maintain HCC cell lines in DMEM with 10% FBS at 37°C with 5% CO₂.
- Seed cells in appropriate plates 24 hours before transfection to achieve 60-70% confluency.
- Transfect with miR-221 mimic, inhibitor, or appropriate negative controls using transfection reagent according to manufacturer's protocol.
Transfection Efficiency Validation:
- Harvest cells 48-96 hours post-transfection for RNA extraction.
- Perform qRT-PCR to verify miR-221 expression changes using U6 snRNA as endogenous control.
- Confirm target regulation by Western blot for known targets (p27, p57, LIFR).
Functional Assays:
- Cell Viability: Perform MTS assay at 24, 48, 72, and 96 hours post-transfection according to manufacturer's instructions.
- Apoptosis Analysis: Use Annexin V-FITC/PI staining followed by flow cytometry 72 hours post-transfection.
- Cell Cycle Analysis: Fix cells in 70% ethanol, treat with RNase A and PI, then analyze by flow cytometry.
Expected Results: miR-221 inhibitor should reduce cell viability, increase apoptosis, and cause G1/S phase arrest, while miR-221 mimic should produce opposite effects [12].
Protocol 2: Investigating GAS5-miRNA Interactions Through RNA Immunoprecipitation
Objective: To validate direct binding interactions between GAS5 and candidate miRNAs (e.g., miR-182, miR-423-3p).
Materials and Reagents:
- Anti-AGO2 antibody (Millipore) and control IgG
- Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore)
- HCC cell lines (SMMC-7721, Hep3B)
- Plasmid constructs for GAS5 overexpression
- TRIzol reagent for RNA extraction
- qRT-PCR equipment and reagents
Procedure:
- Cell Preparation and Transfection:
- Culture HCC cells and transfect with miRNA mimics (miR-182, miR-423-3p, or negative control) using appropriate transfection reagent.
- Incubate for 48 hours to allow miRNA incorporation into RISC complexes.
RNA Immunoprecipitation:
- Lysc cells using RIP lysis buffer containing protease and RNase inhibitors.
- Incubate cell lysates with magnetic beads conjugated with anti-AGO2 antibody or control IgG overnight at 4°C.
- Wash beads extensively with RIP wash buffer.
- Isolate co-precipitated RNA using proteinase K digestion and phenol-chloroform extraction.
Analysis of Precipitated RNA:
- Reverse transcribe purified RNA using random primers.
- Perform qPCR analysis using GAS5-specific primers.
- Calculate enrichment of GAS5 in anti-AGO2 samples compared to IgG controls.
Expected Results: Significant enrichment of GAS5 in anti-AGO2 immunoprecipitates from cells transfected with targeting miRNAs indicates direct binding, supporting the ceRNA mechanism [15] [16].
Signaling Pathway Visualization
Diagram 1: Oncogenic ncRNA Networks in HCC. The illustration shows two key oncogenic mechanisms: GAS5 stabilization through METTL3-mediated m6A modification and subsequent sponging of tumor-suppressive miR-423-3p, leading to SMARCA4-driven oncogenesis; and miR-221-mediated suppression of tumor suppressors p27/p57 and LIFR, promoting proliferation and metastasis [15] [12] [13].
Diagram 2: Tumor-Suppressive Functions and Therapeutic Applications of GAS5. The diagram illustrates GAS5's tumor-suppressive role through sponging oncogenic miR-182 and derepressing ANGPTL1, ultimately inhibiting metastasis. The therapeutic application shows UTMD-mediated GAS5 delivery as a potential treatment approach for HCC [16].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for ncRNA Functional Studies in HCC
| Reagent/Category | Specific Examples | Application Purpose | Experimental Context |
|---|---|---|---|
| Cell Lines | Hep3B, HepG2, SNU449, SMMC-7721, PLC/PRF/5 | In vitro functional studies | Proliferation, apoptosis, migration assays [12] [16] |
| miRNA Modulators | miR-221 mimic/inhibitor, miR-182 mimic, miR-423-3p mimic | Gain/loss-of-function studies | Functional validation of miRNA targets [12] [16] |
| LncRNA Tools | GAS5 overexpression vectors, siGAS5 | Manipulating lncRNA expression | Studying GAS5 functions [15] [16] |
| Specialized Kits | Magna RIP Kit, Dual-Luciferase Reporter Assay, Transfection Reagents | Mechanistic studies | Validating miRNA-ncRNA interactions [15] [16] |
| Animal Models | Ras-transgenic spontaneous HCC mice, Xenograft models | In vivo validation | Therapeutic efficacy studies [15] |
| Therapeutic Delivery | Ultrasound targeted microbubble destruction (UTMD) | Targeted therapy approach | GAS5 delivery for metastasis inhibition [16] |
| Fgi-106 | Fgi-106, CAS:1149348-10-6, MF:C28H42Cl4N6, MW:604.5 g/mol | Chemical Reagent | Bench Chemicals |
| Taletrectinib | Taletrectinib, CAS:1505514-27-1, MF:C23H24FN5O, MW:405.5 g/mol | Chemical Reagent | Bench Chemicals |
The investigation of dysregulated ncRNAs in HCC reveals a complex regulatory network where molecules like miR-221 and GAS5 play critical roles in tumor progression. While miR-221 consistently demonstrates oncogenic properties, GAS5 exhibits context-dependent functions, highlighting the importance of comprehensive functional validation in specific cellular contexts. The experimental protocols and analytical frameworks presented here provide researchers with standardized methodologies for exploring ncRNA functions in HCC, facilitating the identification of novel therapeutic targets. As RNA sequencing technologies continue to evolve, integrating multi-omics data will be essential for unraveling the intricate ncRNA regulatory networks in hepatocellular carcinoma, ultimately leading to improved diagnostic and therapeutic strategies for this devastating disease.
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related death worldwide, with its molecular pathogenesis intricately linked to the dysregulation of key signaling pathways [17]. Next-generation sequencing technologies, particularly RNA-sequencing (RNA-Seq), have revolutionized our understanding of cancer biology by revealing that the vast majority of the human genome is transcribed into non-coding RNAs (ncRNAs) [17] [18]. These ncRNAs, once considered "transcriptional noise," are now recognized as pervasive regulators of essentially all cancer hallmarks, including proliferation, apoptosis, invasion, and metastasis [19]. In the context of a broader thesis on RNA sequencing analysis of ncRNAs in HCC tissues, this application note provides a detailed mechanistic and protocol-oriented overview of how ncRNAs regulate the Wnt/β-catenin and PI3K/AKT pathways—two signaling cascades frequently aberrantly activated in HCC.
The ncRNA Landscape: Categories and Functions
Non-coding RNAs are broadly categorized by size and function. The most studied classes in oncology are microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) [18] [19]. Their biogenesis, structure, and primary mechanisms of action are distinct, as summarized in the table below.
Table 1: Major Classes of Non-Coding RNAs in Cancer
| ncRNA Class | Size | Structure | Primary Functions | Role in Gene Regulation |
|---|---|---|---|---|
| MicroRNA (miRNA) | ~22 nucleotides [18] | Short, single-stranded [18] | Binds mRNA; induces degradation or translational inhibition [18] [20] | Post-transcriptional regulation [20] |
| Long Non-Coding RNA (lncRNA) | >200 nucleotides [17] [18] | Linear, can form complex structures [19] | Guide, decoy, scaffold, or signal for transcription and epigenetic regulation [17] | Epigenetic, transcriptional, and post-transcriptional regulation [17] |
| Circular RNA (circRNA) | >200 nucleotides [18] | Covalently closed loop [19] | Acts as miRNA "sponge," interacts with proteins, can encode peptides [19] | Mainly post-transcriptional regulation [18] |
The subcellular localization of ncRNAs is a critical determinant of their function. Nuclear-enriched lncRNAs, for instance, often regulate transcription and epigenetic modifications, while cytoplasmic lncRNAs more frequently influence mRNA stability and translation [17]. This spatial organization is a key consideration when designing experiments to investigate ncRNA function.
Regulation of the Wnt/β-Catenin Pathway by ncRNAs
The Wnt/β-catenin pathway is a critical regulator of cell fate, proliferation, and stemness. In the "off" state, a destruction complex—comprising AXIN1, APC, CK1, and GSK3β—phosphorylates β-catenin, targeting it for ubiquitination and proteasomal degradation. Upon Wnt ligand binding to Frizzled and LRP5/6 receptors, the destruction complex is disrupted. This allows β-catenin to accumulate in the cytoplasm and translocate to the nucleus, where it partners with TCF/LEF transcription factors to activate target genes (e.g., c-MYC, CYCLIN D1) [20]. Aberrant activation of this pathway is a hallmark of HCC, driving tumor initiation and progression.
Mechanistic Insights into ncRNA-Mediated Regulation
ncRNAs intricately control the Wnt/β-catenin pathway at multiple levels. They can function as either oncogenic drivers or tumor suppressors.
- Oncogenic ncRNAs: Many ncRNAs promote pathway activation. For example, the lncRNA lncTCF7 is highly expressed in liver cancer stem cells (CSCs). It recruits the SWI/SNF chromatin-remodeling complex to the promoter of TCF7, thereby activating the Wnt pathway and sustaining liver CSC self-renewal [18]. Similarly, some miRNAs, like miR-629, can directly target and inhibit negative regulators of the pathway, leading to its hyperactivation [18].
- Tumor-Suppressive ncRNAs: Conversely, other ncRNAs act as pathway brakes. miR-34a, frequently downregulated in cancers, can target and inhibit key components of the Wnt pathway, thereby suppressing cancer stem cell self-renewal [18]. The lncRNA GAS5 is another tumor suppressor, and its low expression in HCC is associated with poor prognosis [8].
Table 2: Key ncRNAs Regulating the Wnt/β-catenin Pathway in HCC
| ncRNA | Type | Expression in HCC | Molecular Target/Mechanism | Functional Outcome |
|---|---|---|---|---|
| lncTCF7 | lncRNA | Upregulated [18] | Recruits SWI/SNF to TCF7 promoter [18] | Activates Wnt signaling, sustains CSC self-renewal [18] |
| miR-34a | miRNA | Downregulated [18] | Inhibits Wnt pathway components [18] | Suppresses CSC self-renewal, tumor suppression [18] |
| GAS5 | lncRNA | Downregulated [8] | Activates CHOP and caspase-9 [8] | Inhibits proliferation, induces apoptosis [8] |
| HOTTIP | lncRNA | Upregulated [18] | Epigenetic regulator of hematopoietic genes [18] | Promotes tumorigenesis (context-dependent) [18] |
Regulation of the PI3K/AKT Pathway by ncRNAs
The PI3K/AKT pathway is a potent regulator of cell growth, survival, metabolism, and therapy response. Activation by growth factors leads to PI3K-mediated generation of PIP3, which recruits AKT to the membrane for activation via phosphorylation. AKT then phosphorylates numerous downstream effectors, including mTOR, to drive anabolic processes and inhibit apoptosis. The tumor suppressor PTEN antagonizes this pathway by dephosphorylating PIP3 back to PIP2. Loss of PTEN or mutation of PIK3CA leads to hyperactive PI3K/AKT signaling, a common event in HCC that promotes proliferation, metastasis, and chemoresistance [21] [22].
The ncRNA/PTEN/PI3K/AKT Axis
A central theme in ncRNA-mediated regulation of this pathway is the control of PTEN. Many oncogenic miRNAs (oncomiRs) directly target the PTEN mRNA for degradation, thereby releasing the brake on the pathway [21]. Furthermore, lncRNAs and circRNAs can act as competing endogenous RNAs (ceRNAs) by sponging these miRNAs, thereby indirectly regulating PTEN expression [21]. The ncRNA/PI3K/Akt axis is a crucial determinant of cell proliferation, metastasis, epithelial-mesenchymal transition (EMT), and therapy resistance in human cancers [21].
Table 3: Key ncRNAs Regulating the PI3K/AKT Pathway in HCC
| ncRNA | Type | Expression in HCC | Molecular Target/Mechanism | Functional Outcome |
|---|---|---|---|---|
| OncomiRs (e.g., miR-155) | miRNA | Upregulated [18] | Directly targets PTEN mRNA [21] [18] | Promotes proliferation, tumor growth [18] |
| LINC00152 | lncRNA | Upregulated [8] | Promotes proliferation via CCDN1; high level predicts poor prognosis [8] | Drives cell proliferation [8] |
| UCA1 | lncRNA | Upregulated [8] | Modulates proliferation and apoptosis [8] | Promotes tumor growth [8] |
| Tumor-Suppressive miRNAs (e.g., Let-7) | miRNA | Downregulated [18] | Targets oncogenes like K-RAS [18] | Inhibits proliferation, induces apoptosis [18] |
Cross-Talk and Therapeutic Implications
Pathway Interdependence in HCC
The Wnt/β-catenin and PI3K/AKT pathways do not function in isolation. Significant cross-talk exists between them, creating a robust network that drives oncogenesis. Research by Li et al. demonstrated that constitutive activation of β-catenin alone induces apoptosis in hematopoietic stem cells (HSCs), while loss of PTEN alone leads to transient HSC expansion followed by exhaustion. However, the combination of both β-catenin activation and Pten deletion drives a synergistic expansion of phenotypic long-term HSCs, illustrating powerful cooperation between the two pathways in controlling self-renewal, apoptosis, and differentiation blockade [23]. This cooperation is highly relevant to HCC, where concurrent dysregulation of both pathways is common.
ncRNAs as Therapeutic Targets and Biomarkers
The critical regulatory role of ncRNAs makes them attractive therapeutic targets. Strategies include:
- Antagonizing Oncogenic ncRNAs: Using antisense oligonucleotides (ASOs) or small interfering RNAs (siRNAs) to knock down oncogenic lncRNAs or miRNAs.
- Restoring Tumor-Suppressive ncRNAs: Delivering synthetic tumor-suppressive miRNAs or lncRNAs using viral or nanoparticle-based systems.
Moreover, the high specificity of ncRNA expression patterns makes them excellent biomarker candidates. For instance, a machine learning model integrating plasma levels of four lncRNAs (LINC00152, LINC00853, UCA1, GAS5) with conventional laboratory data achieved 100% sensitivity and 97% specificity in diagnosing HCC, far outperforming individual biomarkers [8]. The ratio of LINC00152 to GAS5 was also a significant prognostic indicator for mortality risk [8].
Experimental Protocols for Investigating ncRNA-Pathway Interactions
Core Protocol: Validating ncRNA-Target Relationships in HCC Models
This protocol outlines a standard workflow for confirming that a candidate ncRNA regulates a specific signaling pathway in the context of HCC.
Objective: To functionally validate the role of a specific ncRNA (e.g., LINC00152) in modulating the PI3K/AKT pathway in hepatocellular carcinoma cells.
Materials and Reagents:
- Cell Line: Human HCC cell line (e.g., HepG2, Huh-7).
- Culture Medium: DMEM or RPMI-1640 supplemented with 10% FBS and 1% penicillin/streptomycin.
- Transfection Reagents: Lipofectamine RNAiMAX or similar.
- Oligonucleotides:
- siRNA or ASO: Targeting the candidate ncRNA for knockdown.
- Negative Control siRNA/ASO: Scrambled sequence.
- Expression Plasmid: For ncRNA overexpression.
- RNA Isolation Kit: miRNeasy Mini Kit (QIAGEN, 217004) [8].
- cDNA Synthesis Kit: RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622) [8].
- qRT-PCR Reagents: PowerTrack SYBR Green Master Mix (Applied Biosystems, A46012) [8].
- Antibodies: Anti-p-AKT (Ser473), anti-total AKT, anti-PTEN, anti-β-catenin, and corresponding HRP-conjugated secondary antibodies.
- Protein Lysis Buffer: RIPA buffer supplemented with protease and phosphatase inhibitors.
Procedure:
- Cell Seeding and Transfection:
- Seed HCC cells in 6-well or 12-well plates to reach 60-70% confluency at the time of transfection.
- For knockdown, transfect cells with 50 nM of specific siRNA/ASO or negative control using the transfection reagent per manufacturer's instructions.
- For overexpression, transfect with 1-2 µg of plasmid DNA or corresponding empty vector control.
- Incubate for 48-72 hours before analysis.
RNA Isolation and Quantitative RT-PCR (qRT-PCR):
- Extract total RNA using the miRNeasy Mini Kit. Include a DNase digestion step to remove genomic DNA contamination.
- Synthesize cDNA using the RevertAid Kit.
- Perform qRT-PCR using SYBR Green Master Mix on a real-time PCR system. Use GAPDH as a housekeeping gene for normalization [8].
- Analyze data using the ∆∆Ct method to determine relative expression changes of the target ncRNA and potential downstream mRNA targets (e.g., CCND1, MYC).
Protein Extraction and Western Blotting:
- Lyse transfected cells in ice-cold RIPA buffer. Quantify protein concentration using a BCA assay.
- Separate 20-30 µg of total protein by SDS-PAGE and transfer to a PVDF membrane.
- Block the membrane with 5% non-fat milk for 1 hour.
- Incubate with primary antibodies (e.g., 1:1000 dilution for p-AKT, total AKT, PTEN) overnight at 4°C.
- Incubate with HRP-conjugated secondary antibody (1:5000) for 1 hour at room temperature.
- Develop the blot using a chemiluminescent substrate and image. Assess changes in pathway activation (e.g., p-AKT/total AKT ratio, PTEN protein levels, β-catenin levels).
Expected Outcomes: Knockdown of an oncogenic ncRNA (e.g., LINC00152) should result in decreased expression of pathway targets, reduced levels of p-AKT, and potentially increased PTEN protein. Overexpression should have the opposite effect, confirming the ncRNA's role as a pathway activator.
Advanced Protocol: RNA Sequencing for Pathway Discovery
For an unbiased discovery of ncRNAs linked to pathways in HCC tissues, RNA-Seq is the gold standard.
Workflow:
- Sample Preparation: Total RNA is extracted from paired HCC and adjacent non-tumor liver tissues. RNA integrity (RIN > 8.0) is critical.
- Library Preparation: Use kits that preserve all RNA species (including small RNAs for miRNA sequencing) or perform ribosomal RNA depletion to enrich for lncRNAs and circRNAs.
- Sequencing: Perform high-throughput sequencing on a platform such as Illumina NovaSeq to a sufficient depth (e.g., 50-100 million paired-end reads per sample).
- Bioinformatic Analysis:
- Alignment and Quantification: Map reads to the human reference genome (e.g., GRCh38) using aligners like STAR. Quantify expression of known genes and ncRNAs using tools like FeatureCounts.
- Differential Expression: Identify significantly dysregulated ncRNAs between tumor and normal groups using packages like DESeq2 or edgeR.
- Pathway Enrichment Analysis: Input lists of dysregulated ncRNAs and/or their co-expressed protein-coding genes into tools like GSEA or DAVID to identify enriched pathways like "Wnt signaling" or "PI3K-AKT signaling" [24].
- ceRNA Network Construction: Predict miRNA-mRNA and miRNA-lncRNA interactions using databases (TargetScan, miRanda, StarBase) and build competing endogenous RNA networks to visualize how lncRNAs might sponge miRNAs to regulate key pathways.
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Research Reagent Solutions for Investigating ncRNAs in Signaling Pathways
| Reagent / Tool Category | Specific Examples | Function / Application |
|---|---|---|
| RNA Isolation & QC | miRNeasy Mini Kit (QIAGEN) [8] | Simultaneous purification of total RNA, including small RNAs, essential for ncRNA studies. |
| cDNA Synthesis | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) [8] | High-efficiency reverse transcription for subsequent qRT-PCR analysis of ncRNAs and mRNAs. |
| qRT-PCR Analysis | PowerTrack SYBR Green Master Mix (Applied Biosystems) [8] | Sensitive and specific detection for quantifying ncRNA expression levels. |
| Functional Modulation | Silencer Select Pre-designed siRNAs/ASOs (Thermo Fisher); LNA GapmeRs (Exiqon) | Tools for efficient knockdown of nuclear or cytoplasmic ncRNAs. |
| Pathway Activity Assays | Phospho-AKT (Ser473) Antibody (CST); β-Catenin Antibody (BD Biosciences) | Key reagents for Western Blot to measure pathway activity upon ncRNA manipulation. |
| Bioinformatics Databases | miRBase; lncRNAdb; StarBase; TargetScan | Curated resources for ncRNA annotation, target prediction, and interaction validation. |
| Columbamine chloride | Columbamine chloride, CAS:1916-10-5, MF:C20H20ClNO4, MW:373.8 g/mol | Chemical Reagent |
| Ganfeborole | Ganfeborole, CAS:2131798-12-2, MF:C10H13BClNO4, MW:257.48 g/mol | Chemical Reagent |
The intricate regulation of the Wnt/β-catenin and PI3K/AKT pathways by ncRNAs represents a fundamental layer of control in HCC pathogenesis. RNA sequencing studies of HCC tissues continue to uncover novel ncRNAs and their complex networks. The experimental protocols and tools detailed herein provide a roadmap for researchers to validate these interactions and explore their therapeutic potential. Targeting specific ncRNAs, or leveraging them as biomarkers in sophisticated diagnostic panels, holds immense promise for improving the prognosis of HCC patients.
Hepatocellular carcinoma (HCC) constitutes approximately 90% of primary liver cancers and ranks as the third leading cause of cancer-related deaths globally [25] [26]. The molecular pathogenesis of HCC involves complex biological processes, including DNA damage, epigenetic modifications, and oncogene mutations [27] [28]. Over the past decade, non-coding RNAs (ncRNAs) have emerged as critical regulators of gene expression, playing pivotal roles in HCC progression despite lacking protein-coding capacity [29] [30]. The dysregulation of ncRNAs, including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), contributes significantly to fundamental cancer hallmarks such as sustained proliferation, metastasis, and angiogenesis [25] [26]. This application note details the mechanistic links between ncRNA dysregulation and these HCC hallmarks, providing experimental protocols and analytical frameworks for researchers investigating ncRNA functions in hepatocarcinogenesis within the broader context of RNA sequencing analysis of non-coding RNAs in HCC tissues.
Quantitative Profiling of Prognostic ncRNAs in HCC
Comprehensive analyses of HCC tissue specimens have identified numerous ncRNAs with significant prognostic value, highlighting their clinical relevance as biomarkers and therapeutic targets.
Table 1: Prognostic lncRNAs in Hepatocellular Carcinoma
| LncRNA Name | Expression in HCC | Biological Function | Prognostic Value (HR [95% CI]) | Reference |
|---|---|---|---|---|
| LINC00152 | Upregulated | Promotes cell proliferation | Shorter OS: HR 2.524 [1.661-4.015] | [31] |
| LINC01554 | Downregulated | Suppresses tumor growth | Shorter OS: HR 2.507 [1.153-2.832] | [31] |
| HOXC13-AS | Upregulated | Enhances invasion | Shorter OS: HR 2.894 [1.183-4.223] | [31] |
| LASP1-AS | Downregulated | Inhibits metastasis | Shorter OS: HR 3.539 [2.698-6.030] | [31] |
| ELMO1-AS1 | Upregulated | Tumor suppressor | Longer OS: HR 0.430 [0.225-0.824] | [31] |
| GAS5-AS1 | Upregulated | Tumor suppressor | Longer OS: HR 0.370 [0.153-0.898] | [31] |
Table 2: Dysregulated miRNAs in HBV-Related HCC and Their Functional Roles
| miRNA | Expression | Role in HCC | Target Genes/Pathways | Reference |
|---|---|---|---|---|
| miR-17-5p | Upregulated | Oncogenic | HIF1A, Myc (stemness maintenance) | [32] |
| miR-21 | Upregulated | Oncogenic | PDCD4, PTEN | [30] |
| miR-221/222 | Upregulated | Oncogenic | CXCL4/12, TFRC | [30] |
| miR-122 | Downregulated | Tumor suppressor | PKM2, SLC7A1 (metabolism) | [30] |
| miR-199a/b | Downregulated | Tumor suppressor | ROCK1, PI3K/Akt | [30] |
| miR-125b | Downregulated | Tumor suppressor | VEGFA, cyclin D2/E2 | [30] |
Molecular Mechanisms Linking ncRNA Dysregulation to HCC Hallmarks
Sustained Proliferation and Evasion of Growth Suppression
ncRNAs orchestrate hepatocellular proliferation through intricate regulation of core signaling pathways and cell cycle components. The AURKA kinase represents a critical node in proliferation control, with its expression modulated by multiple ncRNAs [25]. In HCC, lncRNA H19 stimulates proliferation by downregulating miRNA-15b expression and activating the CDC42/PAK1 axis [28]. Similarly, lncRNA-p21 forms a positive feedback loop with HIF-1α to drive glycolysis, thereby supporting tumor growth under hypoxic conditions [28]. The miR-17-92 cluster, frequently upregulated in HBV-related HCC, promotes proliferation by targeting estrogen receptor alpha and components of the cell cycle machinery [30]. Cancer stem cells (CSCs), responsible for tumor initiation and therapy resistance, are maintained by ncRNAs such as miR-17-5p, which preserves stemness properties by targeting HIF1A and Myc [32].
Activation of Invasion and Metastasis
Metastatic progression in HCC is driven by ncRNA-mediated regulation of epithelial-mesenchymal transition (EMT), cytoskeletal reorganization, and extracellular matrix remodeling. AURKA overexpression promotes EMT through the PI3K/AKT and MAPK pathways, increasing expression of N-cadherin and CSC markers (CD133, CD44) [25]. The lncRNA NEAT1 facilitates HCC cell migration and invasion through diverse mechanisms, including interaction with miRNAs and proteins [27]. In HBV-related HCC, miR-30a-5p downregulation enhances EMT by losing repression of SNAIL1, a key transcriptional regulator of EMT [30]. Additionally, lncRNAs such as DSCR8, PNUTS, and HULC contribute to migration and apoptosis resistance through distinct molecular mechanisms [27].
Induction of Angiogenesis
Angiogenesis represents a hallmark of HCC, supported by ncRNA-mediated regulation of pro-angiogenic factors. The VEGF/VEGFR pathway is particularly important in HCC, a highly vascular tumor, with VEGFA demonstrating 7-14% frequency of focal amplification in HCC [26]. The miR-17-92 cluster promotes angiogenesis in HBV-related HCC, facilitating tumor vascularization [30]. Conversely, tumor-suppressive miR-125b inhibits angiogenesis by targeting VEGFA, with its downregulation in HCC contributing to enhanced vascularization [30]. The efficacy of VEGFR-targeted therapies in HCC, including bevacizumab and ramucirumab, underscores the clinical relevance of angiogenesis in HCC management [26].
Therapeutic Resistance and Autophagy
The lncRNA-autophagy axis represents a crucial mechanism of therapeutic resistance in HCC. Autophagy plays a paradoxical role in hepatocarcinogenesis, acting as a tumor suppressor during initiation but promoting survival and progression in advanced stages [33]. LncRNAs regulate key autophagy signaling networks (e.g., PI3K/AKT/mTOR, AMPK, Beclin-1) and modulate resistance to first-line agents by altering autophagic flux [33]. In hypoxic conditions, linc-RoR functions as a miR-145 sponge, upregulating p70S6K1, PDK1, and HIF-1α to accelerate proliferation and potentially contribute to therapy resistance [28].
Diagram 1: ncRNA Dysregulation in HCC Hallmarks. This diagram illustrates the central role of ncRNA dysregulation in driving key hepatocellular carcinoma hallmarks through multiple molecular pathways, ultimately leading to adverse clinical outcomes.
Experimental Protocols for ncRNA Functional Analysis
Protocol: Identification of HCC-Associated ncRNAs from RNA Sequencing Data
Purpose: To identify differentially expressed ncRNAs in HCC tissues compared to non-tumor liver tissues using RNA sequencing data.
Materials and Reagents:
- RNA extraction kit (e.g., TRIzol)
- RNA quality control equipment (Bioanalyzer)
- RNA sequencing library preparation kit
- High-throughput sequencer (Illumina)
- Computational resources for bioinformatics analysis
Procedure:
- Tissue Collection: Obtain paired HCC and adjacent non-tumor liver tissues from patients undergoing surgical resection (ethical approval required).
- RNA Extraction: Isolate total RNA using TRIzol reagent according to manufacturer's protocol.
- RNA Quality Control: Assess RNA integrity using Bioanalyzer (RIN >7.0 required).
- Library Preparation: Prepare RNA sequencing libraries using strand-specific protocols to preserve strand orientation information.
- Sequencing: Perform high-throughput sequencing (minimum 50 million paired-end reads per sample).
- Bioinformatics Analysis:
- Align reads to reference genome (GRCh38) using splice-aware aligners (STAR or HISAT2).
- Assemble transcripts and quantify expression using StringTie or Cufflinks.
- Identify differentially expressed ncRNAs using DESeq2 or edgeR (FDR <0.05, |log2FC| >1).
- Validate findings using public datasets (TCGA-LIHC, GEO).
Troubleshooting Tip: For ncRNA quantification, use specialized annotation databases (LNCipedia, NONCODE) in addition to standard references to ensure comprehensive ncRNA coverage.
Protocol: Functional Validation of ncRNAs in HCC Cell Models
Purpose: To validate the functional role of candidate ncRNAs in HCC proliferation, migration, and angiogenesis using in vitro models.
Materials and Reagents:
- HCC cell lines (Huh7, HepG2, Hep3B)
- Culture media and supplements
- Lipofectamine RNAiMAX transfection reagent
- ncRNA mimics/inhibitors or siRNA/shRNA constructs
- qRT-PCR reagents
- Cell proliferation assay kit (CCK-8/MTS)
- Migration assay equipment (Transwell chambers)
- Tube formation assay materials (Matrigel, HUVEC cells)
Procedure:
- Cell Culture: Maintain HCC cell lines in recommended media under standard conditions (37°C, 5% CO2).
- ncRNA Modulation:
- For gain-of-function: Transfert with ncRNA mimics or expression vectors.
- For loss-of-function: Transfert with ncRNA inhibitors or siRNA/shRNA.
- Include appropriate negative controls (scrambled sequences).
- Efficiency Validation: Confirm modulation efficiency by qRT-PCR after 24-48 hours.
- Functional Assays:
- Proliferation: Seed transfected cells in 96-well plates and measure viability at 0, 24, 48, and 72h using CCK-8 assay.
- Migration: Perform Transwell migration assay - count migrated cells after 24h.
- Angiogenesis: Collect conditioned media from transfected cells and apply to HUVEC cells on Matrigel - quantify tube formation after 6h.
- Mechanistic Studies:
- Analyze pathway activation (Western blot for PI3K/AKT, MAPK, AURKA).
- Identify direct targets using RIP-seq or CLIP-seq for lncRNAs.
- Validate miRNA targets using luciferase reporter assays.
Troubleshooting Tip: Include rescue experiments by co-transfecting ncRNA modulators with their validated target genes to confirm specificity of observed phenotypes.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Research Reagent Solutions for ncRNA Studies in HCC
| Reagent/Category | Specific Examples | Research Application | Key Considerations |
|---|---|---|---|
| HCC Cell Models | Huh7, HepG2, Hep3B, PLC/PRF/5 | In vitro functional studies | Select based on genetic background; Huh7 supports CSC culture [32] |
| CSC Culture | Ultra-low attachment plates, defined media | Cancer stem cell studies | Enables sphere formation and stemness maintenance [32] |
| ncRNA Modulation | miRNA mimics/inhibitors, siRNA, shRNA, CRISPR/Cas9 | Functional validation | Include appropriate negative controls; optimize delivery efficiency |
| Detection Methods | qRT-PCR, RNAscope, Northern blot, RNA-seq | ncRNA expression quantification | qRT-PCR requires stem-loop primers for miRNAs; RNAscope for spatial resolution |
| Delivery Systems | Lipofectamine, exosomes, chitosan nanoparticles | Therapeutic targeting | Natural nanoparticles (exosomes, chitosan) enhance delivery efficiency [34] |
| Pathway Reporters | Luciferase constructs, GFP-tagged proteins | Mechanism elucidation | Validate direct interactions (e.g., miRNA-mRNA) |
| Animal Models | PDX, xenograft, genetically engineered mice | In vivo validation | Consider microenvironment influences on ncRNA function |
| Lenacapavir | Lenacapavir | Research-grade Lenacapavir, a first-in-class HIV-1 capsid inhibitor. For Research Use Only. Not for human consumption. | Bench Chemicals |
| Regorafenib Monohydrate | Regorafenib Monohydrate, CAS:1019206-88-2, MF:C21H17ClF4N4O4, MW:500.8 g/mol | Chemical Reagent | Bench Chemicals |
The comprehensive integration of ncRNA profiling with functional validation provides powerful insights into HCC pathogenesis and reveals novel therapeutic opportunities. The dysregulation of specific ncRNAs, including H19, NEAT1, miR-17-5p, and miR-122, contributes fundamentally to HCC hallmarks through identifiable molecular mechanisms. The experimental frameworks outlined herein enable researchers to systematically investigate these relationships, from initial discovery through mechanistic validation. As research advances, targeting ncRNA networks holds promise for developing innovative diagnostic biomarkers and therapeutic strategies to improve outcomes for HCC patients. The continued integration of multi-omics approaches will be essential for validating these candidates and translating ncRNA research into clinical applications.
The Emerging Role of ncRNAs in Modulating the Tumor Immune Microenvironment
Hepatocellular carcinoma (HCC) represents a major global health challenge characterized by a complex tumor immune microenvironment (TIME) that plays a pivotal role in tumor progression and therapeutic response [35]. Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), have emerged as critical regulators of gene expression and immune cell function within the HCC landscape [35] [36]. These molecules account for approximately 98% of the transcribed genome and demonstrate significant dysregulation in HCC, affecting various biological processes from immune evasion to therapy resistance [36].
The immunosuppressive nature of the HCC microenvironment presents a substantial barrier to effective treatment, particularly for immunotherapies such as immune checkpoint inhibitors (ICIs) [35]. ncRNAs have been shown to directly influence this immunosuppression by regulating the infiltration and activation of immune cells, shaping cytokine profiles, and controlling immune checkpoint expression [35] [37]. Understanding these regulatory mechanisms provides crucial insights for developing novel diagnostic biomarkers and therapeutic strategies aimed at reprogramming the TIME to enhance anti-tumor immunity.
Table 1: Major ncRNA Classes and Their Characteristics in HCC
| ncRNA Class | Size | Key Characteristics | Primary Functions |
|---|---|---|---|
| miRNAs | ~22 nt | Endogenous transcripts; most abundant studied ncRNAs | Regulate ~30% of human genes by binding to 3'UTR of target mRNAs [36] |
| lncRNAs | >200 nt | High tissue and temporal specificity; diverse modes of action | Act as signals, decoys, scaffolds, or guides; regulate transcription and post-transcriptional processes [35] [28] |
| circRNAs | Variable | Closed-loop structure; high stability and conservation | Function as miRNA sponges, bind RBPs, translate peptides, regulate transcription [36] |
ncRNA Regulation of Immune Cells in HCC
T Cell Modulation
T cells are crucial mediators of anti-tumor immunity, and ncRNAs extensively regulate their function within the HCC microenvironment. CD8+ T cells, key effectors in anti-tumor responses, experience functional exhaustion in HCC, characterized by increased expression of inhibitory receptors like PD-1, TIM-3, and LAG-3 [36]. The lncRNA NEAT1 demonstrates significant upregulation in peripheral blood mononuclear cells (PBMCs) of HCC patients and contributes to T-cell exhaustion by binding to miR-155 and regulating the miR-155/Tim-3 pathway. Downregulation of NEAT1 inhibits CD8+ T cell apoptosis and enhances their cytolytic activity against HCC cells, identifying it as a potential target for immunotherapy enhancement [35] [36].
Lnc-Tim3 represents another critical regulator expressed highly in tumor-infiltrating CD8+ T cells. This lncRNA specifically binds to Tim-3 and blocks its interaction with Bat3, thereby inhibiting downstream Lck/NFAT1/AP-1 signaling and exacerbating CD8+ T lymphocyte exhaustion [36]. Targeting Lnc-Tim3 may therefore reverse T-cell dysfunction and improve anti-tumor immunity.
CD4+ T cell differentiation and function are similarly regulated by ncRNAs. These cells can differentiate into various helper T cell subsets (Th1, Th2, Th17) or immunosuppressive regulatory T cells (Tregs), with ncRNAs influencing this differentiation process through complex regulatory networks [36].
Myeloid Cell Regulation
Myeloid cells, including macrophages, dendritic cells (DCs), and myeloid-derived suppressor cells (MDSCs), play diverse roles in the HCC immune landscape. Tumor-associated macrophages (TAMs) often exhibit an M2-polarized, pro-tumor phenotype promoted by specific ncRNAs. Similarly, DC dysfunction impairs antigen presentation and T cell activation, while MDSCs directly suppress T cell responses [35] [38].
ncRNAs can modulate the recruitment and polarization of these myeloid populations through various mechanisms. For instance, certain lncRNAs enhance the recruitment of immunosuppressive cells like MDSCs and Tregs, thereby promoting an environment conducive to tumor growth [35]. In contrast, other ncRNAs may support anti-tumor myeloid functions, highlighting the complex and context-dependent nature of ncRNA-mediated regulation.
Table 2: Key ncRNAs Regulating Immune Cells in HCC
| ncRNA | Type | Target/Mechanism | Effect on TIME |
|---|---|---|---|
| NEAT1 | lncRNA | Binds miR-155, regulating Tim-3 expression | Promotes CD8+ T cell apoptosis and exhaustion [35] [36] |
| Lnc-Tim3 | lncRNA | Binds Tim-3, blocking Bat3 interaction | Inhibits Lck/NFAT1/AP-1 signaling, exacerbating T cell exhaustion [36] |
| CircMET | circRNA | miR-30-5p/Snail/DPP4 axis | Reduces CD8+ T cell infiltration; DPP4 inhibition enhances anti-PD1 efficacy [36] |
| Lnc-Tim3 | lncRNA | Tim-3 signaling pathway | Prevents Bat3-Tim-3 interaction, exacerbating CD8+ T cell exhaustion [35] |
ncRNA Regulation of Immune Checkpoints and Cytokines
Immune Checkpoint Control
Immune checkpoint molecules such as PD-1, PD-L1, and CTLA-4 play crucial roles in regulating immune responses in HCC, and their expression is frequently modulated by ncRNAs [35] [37]. The upregulation of PD-L1 on tumor cells has been particularly associated with poor clinical outcomes, enabling cancer cells to evade immune detection [35].
Multiple miRNAs directly target immune checkpoints in HCC. MiR-374b and miR-4717 target PD-1 and are frequently downregulated in liver cancer, thereby contributing to immune evasion [37]. Similarly, circRNAs such as circUHRF1 are upregulated in HCC and promote PD-1 expression, further enhancing immunosuppression [37]. These findings highlight the multi-layered ncRNA regulatory network controlling immune checkpoint expression in HCC.
Cytokine and Chemokine Regulation
The cytokine milieu within the HCC microenvironment significantly influences immune cell behavior and tumor progression. Pro-inflammatory cytokines such as IL-6 and TNF-α often dominate the HCC landscape, promoting tumor proliferation and facilitating immune evasion [35]. These cytokines can enhance the recruitment of immunosuppressive cells while inhibiting the function of effector immune cells.
ncRNAs play crucial roles in shaping this cytokine environment. For instance, certain lncRNAs have been shown to alter cytokine production, thereby influencing the balance between pro-tumor and anti-tumor immunity [35]. A dysregulated cytokine profile can lead to chronic inflammation, which is a hallmark of HCC development and progression, further emphasizing the importance of ncRNA-mediated regulation in maintaining immune homeostasis.
Experimental Protocols for ncRNA-TIME Analysis
Protocol 1: Comprehensive ncRNA-mRNA Interaction Mapping
Purpose: To identify and validate physical interactions between ncRNAs and immune-related mRNAs in HCC.
Materials:
- HCC tissue samples (tumor and adjacent normal)
- TRIzol reagent for RNA extraction
- DNase I for DNA removal
- cDNA synthesis kit
- SYBR Green PCR Master Mix
- Roche real-time PCR system
- LncTAR computational tool
- miRWalk and miRTarBase databases
Procedure:
- Sequence Retrieval: Obtain sequences of target ncRNAs (e.g., lnc-LRR1-1:1, lnc-LRR1-1:2, hsacirc0001380) from specialized databases (LncPedia, CircBank) and mRNA sequences (e.g., LEF1, MOB1A, PRKCB, SMARCA2) from NCBI [11].
- Physical Interaction Prediction: Use LncTAR tool with minimum free energy threshold of -15 kcal/mol to predict putative interactions between ncRNAs and mRNAs based on complementary base pairing and thermodynamic stability [11].
- miRNA Integration: Analyze 3'UTR and 5'UTR regions of selected genes using miRWalk database filtered for miRTarBase-validated miRNAs to identify potential competing endogenous RNA (ceRNA) networks [11].
- Experimental Validation: Perform qPCR analysis in HCC cell lines (e.g., HepG2) and normal controls (e.g., fibroblast NIH) using gene-specific primers and β-actin normalization to validate predicted interactions [11].
Applications: This protocol enables systematic mapping of ncRNA interactions with key immune and Hippo pathway genes in HCC, revealing novel regulatory mechanisms in HCC progression.
Protocol 2: ncRNA-Mediated T Cell Function Analysis
Purpose: To investigate the role of specific ncRNAs in regulating T cell exhaustion and function in HCC.
Materials:
- PBMCs from HCC patients and healthy donors
- CD8+ T cell isolation kit
- NEAT1 and Lnc-Tim3 expression vectors/siRNAs
- Anti-Tim-3 antibodies
- Flow cytometry equipment
- Cytotoxicity assay reagents
- Apoptosis detection kit
Procedure:
- Cell Isolation and Culture: Isolate CD8+ T cells from PBMCs using magnetic bead-based separation. Culture cells in appropriate media with IL-2 supplementation [35] [36].
- ncRNA Modulation: Transfect CD8+ T cells with NEAT1-targeting siRNAs or Lnc-Tim3 expression vectors using appropriate transfection reagents.
- Functional Assays:
- Cytolytic Activity: Co-culture transfected CD8+ T cells with HCC cell lines and measure specific lysis using 51Cr-release or flow cytometry-based cytotoxicity assays.
- Apoptosis Analysis: Assess CD8+ T cell apoptosis using Annexin V/propidium iodide staining and flow cytometry.
- Cytokine Production: Measure IFN-γ and TNF-α production by ELISA after T cell activation.
- Mechanistic Studies:
- Analyze Tim-3 expression by flow cytometry and Western blot.
- Examine downstream signaling (Lck/NFAT1/AP-1) through Western blot and luciferase reporter assays [36].
- Validate miR-155 interaction with NEAT1 using RNA immunoprecipitation.
Applications: This protocol enables detailed investigation of how specific lncRNAs regulate T cell exhaustion in HCC, providing insights for developing combination immunotherapies targeting ncRNA pathways.
Signaling Pathways and Visualization
ncRNA-Mediated Regulation of Immune Checkpoints in HCC
Diagram 1: ncRNA Regulation of Immune Checkpoints in HCC. This diagram illustrates how different classes of ncRNAs regulate key immune checkpoints in hepatocellular carcinoma, contributing to T-cell exhaustion and tumor immune evasion.
ncRNA Cross-talk in HCC Immune Microenvironment
Diagram 2: ncRNA-Mediated Immunosuppression in HCC. This diagram illustrates the complex network of ncRNA-mediated regulation within the hepatocellular carcinoma immune microenvironment, highlighting how dysregulated ncRNAs promote immunosuppression through multiple cellular mechanisms.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for ncRNA-TIME Studies
| Reagent/Category | Specific Examples | Application/Function | Experimental Context |
|---|---|---|---|
| Cell Isolation Kits | CD8+ T cell isolation kit; PBMC separation kits | Immune cell purification for functional studies | Isulating specific immune populations from blood or tissue [36] |
| ncRNA Modulation Tools | NEAT1 siRNAs; Lnc-Tim3 expression vectors; miRNA mimics/inhibitors | Gain/loss-of-function studies | Investigating specific ncRNA roles in immune regulation [35] [36] |
| Detection Assays | Flow cytometry antibodies (anti-Tim-3, anti-PD-1); ELISA kits (IFN-γ, TNF-α); Apoptosis detection kits | Immune phenotype and functional analysis | Measuring immune checkpoint expression, cytokine production, cell death [36] |
| Computational Resources | LncTAR; miRWalk; miRTarBase; GEO database | Interaction prediction and data mining | Predicting ncRNA-mRNA interactions; analyzing expression datasets [11] |
| Cell Culture Models | HepG2 HCC cells; normal fibroblast controls; patient-derived PBMCs | In vitro validation systems | Experimental validation of ncRNA functions in relevant cellular contexts [11] |
Concluding Remarks
The emerging role of ncRNAs in modulating the tumor immune microenvironment of HCC represents a paradigm shift in our understanding of liver cancer biology and therapeutic resistance. These regulatory molecules influence virtually all aspects of the HCC immune landscape, from T cell exhaustion and checkpoint expression to myeloid cell polarization and cytokine signaling. The intricate networks formed by different ncRNA classes highlight the complexity of immune regulation in HCC and underscore the need for comprehensive analytical approaches.
Moving forward, the clinical translation of ncRNA research holds significant promise for improving HCC management. ncRNAs show potential as predictive biomarkers for immunotherapy response and as therapeutic targets themselves [38]. Combining ncRNA-targeting strategies with existing immunotherapies may help overcome current limitations in HCC treatment by reprogramming the immunosuppressive microenvironment. However, challenges remain in delivery specificity, off-target effects, and understanding context-dependent functions, necessitating further research into the precise mechanisms of ncRNA action in the HCC immune ecosystem.
As sequencing technologies advance and multi-omics integration becomes more sophisticated, our ability to decipher the complex ncRNA networks governing HCC immunity will continue to improve, ultimately paving the way for more effective, personalized immunotherapeutic approaches for this devastating malignancy.
From Raw Data to Discovery: Methodological Pipelines for ncRNA Sequencing
Hepatocellular carcinoma (HCC) represents a major global health concern, ranking as the sixth most frequently diagnosed cancer worldwide and the third leading cause of cancer-related deaths [26]. Its complex molecular heterogeneity, characterized by diverse etiologies including hepatitis B (HBV) and C (HCV), metabolic disorders, and environmental factors such as aflatoxin exposure, presents significant challenges for research [26] [39]. The molecular etiology of HCC differs substantially depending on specific etiologies and genotoxic damage, necessitating carefully designed studies to account for this variability [26]. For investigations focusing on RNA sequencing analysis of non-coding RNAs in HCC tissues, rigorous experimental design encompassing tissue acquisition, sample size determination, and quality control is paramount to generating reliable, reproducible data that accurately reflects the disease's complexity.
The development of HCC is typically a multistep process arising from malignant transformation of hepatocytes that acquire diverse genomic and epigenomic alterations [39]. Several signaling pathways are frequently dysregulated in HCC, including Wnt/β-catenin, phosphatidylinositol-3-kinase and protein kinase B, and various receptor tyrosine kinase pathways, leading to uncontrolled cell proliferation, metastasis, and recurrence [26]. Within this complex molecular landscape, long non-coding RNAs (lncRNAs) have emerged as pivotal players in HCC, influencing its initiation, progression, invasion, and metastasis through regulation of gene expression at epigenetic, transcriptional, and post-transcriptional levels [40]. This application note provides a comprehensive framework for designing robust HCC studies focused on RNA sequencing analysis, with particular emphasis on tissue acquisition strategies, sample size calculation, and quality control procedures tailored to non-coding RNA research.
Tissue Acquisition and Annotation Protocols
Patient Cohort Stratification and Ethical Considerations
Proper tissue acquisition begins with careful patient cohort stratification based on clinically relevant parameters. The etiology of HCC significantly influences its molecular characteristics; HBV-associated HCC exhibits distinct molecular subtypes and immune responses compared to NASH-induced HCC [26]. Table 1 outlines essential patient clinical data that should be collected during cohort stratification to ensure sample relevance and enable subsequent data analysis.
Table 1: Essential Patient Clinical Data for HCC Cohort Stratification
| Data Category | Specific Parameters | Research Significance |
|---|---|---|
| Demographics | Age, Gender, Ethnicity | Account for population-specific variations [41] |
| Etiology | HBV, HCV, NAFLD/NASH, Alcohol-related | Different molecular pathways are activated by different etiologies [26] |
| Liver Function | Child-Pugh Stage, MELD Score | Determines degree of liver dysfunction and compensation [41] |
| Tumor Staging | BCLC Stage, TNM Classification | Correlates molecular findings with disease progression [42] |
| Histopathological Features | Edmondson Grade, Tumor Size, Vascular Invasion | Associates molecular data with pathological characteristics [43] [40] |
| Prior Treatments | Surgical Resection, Locoregional Therapies, Systemic Treatments | Affects tissue molecular landscape [41] |
Ethical considerations must be addressed prior to tissue collection. Institutional Review Board approval and informed patient consent are mandatory, with specific provisions for biospecimen collection, storage, and future use [41]. Documentation should include consent for longitudinal sample collection where applicable, particularly for studies investigating disease progression or treatment response.
Tissue Collection and Processing Workflow
A standardized protocol for tissue collection and processing is essential to maintain RNA integrity, particularly for non-coding RNA studies. The workflow should be optimized to minimize ischemic time and preserve RNA quality. The following protocol outlines key steps for tissue acquisition:
Protocol: HCC Tissue Collection and Processing for RNA Sequencing
Pre-collection Preparation:
- Coordinate with surgical team to minimize ischemic time
- Prepare sterile containers with appropriate RNA stabilization solution (e.g., RNAlater)
- Label all containers with unique patient identifiers
- Cool transport media to 4°C
Intraoperative Collection:
- Record exact ischemic time (time from devascularization to tissue freezing)
- Collect matched tissue samples from:
- Tumor tissue (avoid necrotic areas)
- Adjacent non-tumor liver tissue (≥2 cm from tumor margin)
- When available, portal vein tumor thrombus (PVTT) or metastatic lesions [44]
- Multiple regions of large tumors should be sampled to account for intra-tumoral heterogeneity
Tissue Processing:
- Divide tissue samples into aliquots for:
- RNA extraction (snap-freeze in liquid nitrogen)
- Formalin-fixed paraffin-embedding (FFPE)
- Fresh tissue preservation (if required for additional assays)
- Snap-freezing should occur within 30 minutes of resection
- Record sample weights for normalization purposes
- Divide tissue samples into aliquots for:
Quality Assessment:
- Perform rapid frozen section confirmation of tissue type
- Document percentage of tumor cellularity
- Note presence of fibrosis, inflammation, or steatosis
Storage:
- Store RNA samples at -80°C
- Maintain detailed inventory with cross-referenced clinical data
This comprehensive approach to tissue acquisition ensures that samples are properly characterized and preserved for subsequent RNA sequencing analysis, particularly important for lncRNA studies which require high-quality RNA.
Sample Size Determination for HCC Studies
Statistical Foundations and Considerations
Appropriate sample size calculation is fundamental to ensuring sufficient statistical power to detect meaningful biological differences in HCC studies. The sample size depends on several factors, including the type of study, α (type I error) and β (type II error) values, effect size, and variability in the data [45]. For HCC research specifically, additional considerations include disease heterogeneity, etiological factors, and tumor staging.
The following protocol provides a framework for calculating sample sizes in HCC studies:
Protocol: Sample Size Calculation for HCC Transcriptomic Studies
Define Primary Objectives:
- Clearly state the main hypothesis to be tested
- Identify primary and secondary endpoints
- For multiple primary objectives, calculate sample size for each and select the largest [45]
Establish Statistical Parameters:
- Set α value (typically 0.05) and β value (typically 0.10 or 0.20) [45]
- Determine statistical power (1-β, typically 80% or 90%)
- Specify allocation ratio if multiple groups are compared
Estimate Effect Size:
- Obtain from pilot data, previous studies, or literature
- For HCC grading studies, an effect size of 1.6 may be appropriate, corresponding to a difference of about 10 units in attenuation values between poorly differentiated and non-poorly differentiated HCC [43]
- Alternatively, a more conservative effect size of 1.0 may be used, corresponding to a difference of about 6 units [43]
Calculate Sample Size:
- Use appropriate formula or statistical software
- G*Power 3.1.9.4 software is recommended for various statistical tests [43]
- For comparative studies of HCC grading with α=0.05, power=0.8, and allocation ratio of 1:3, a total of 18 lesions (14 NP-HCC lesions and 4 P-HCC lesions) would be sufficient to achieve an effect size of 1.6 [43]
- For a more conservative effect size of 1.0 under the same parameters, a total of 52 lesions would be required [43]
Account for Attrition:
- Increase calculated sample size by 10-15% to accommodate potential sample exclusion due to quality control failures
Table 2: Sample Size Requirements for HCC Studies Based on Different Parameters
| Study Design | Effect Size | Power | α | Allocation Ratio | Total Sample Required | Group Distribution |
|---|---|---|---|---|---|---|
| HCC Grading Comparison [43] | 1.6 | 80% | 0.05 | 1:3 | 18 | 14 NP-HCC, 4 P-HCC |
| HCC Grading Comparison [43] | 1.0 | 80% | 0.05 | 1:3 | 52 | 36 NP-HCC, 16 P-HCC |
| Prospective Cohort [41] | N/A | N/A | N/A | N/A | 1600 | 800 per country |
The prospective STOP HCC study exemplifies large-scale sample size planning, recruiting 1,600 patients with advanced fibrosis or compensated cirrhosis to validate the GALAD score for early HCC detection [41]. This sample size provides sufficient power for a phase IV biomarker validation study across multiple clinical sites.
Special Considerations for Single-Cell RNA Sequencing Studies
Single-cell RNA sequencing (scRNA-seq) presents unique sample size considerations due to its high resolution and capacity to capture tumor heterogeneity. The number of patients and cells required depends on the research question and expected cellular diversity.
Protocol: Sample Size Determination for scRNA-seq in HCC
Patient Cohort:
- Include multiple patients (typically 8-12) to account for inter-patient heterogeneity [44]
- Stratify patients based on etiology, tumor stage, and other relevant clinical parameters
Cell Number Calculation:
- Aim for 5,000-10,000 high-quality cells per sample after quality control [44]
- Process more cells initially to account for QC exclusion (approximately 20-30% attrition)
Quality Control Metrics:
For example, a scRNA-seq study of HCC analyzed data from 10 HCC patients from four sites including primary tumor, portal vein tumor thrombus, metastatic lymph node, and non-tumor liver tissue [44]. After quality control filtering using criteria of 4000 > nFeature_RNA > 500 and percent.mt < 10, the number of cells retained ranged from 2,568 to 10,644 across samples [46].
Quality Control Procedures for HCC RNA Sequencing
RNA Quality Assessment and Library Preparation
Quality control is particularly critical for non-coding RNA studies in HCC due to the potential for degradation and the diverse molecular subtypes present in HCC tissues. The following protocol outlines comprehensive QC procedures:
Protocol: Quality Control for HCC RNA Sequencing Studies
RNA Extraction and Quality Assessment:
- Use standardized RNA extraction kits with DNase treatment
- Assess RNA integrity using Agilent Bioanalyzer or TapeStation
- Require minimum RNA Integrity Number (RIN) of ≥7.0 for bulk RNA-seq
- Require RIN ≥8.0 for single-cell RNA-seq applications
- Quantify RNA using fluorometric methods (e.g., Qubit)
Library Preparation and QC:
- Use ribosomal RNA depletion rather than poly-A selection to capture non-coding RNAs
- Validate library size distribution using Bioanalyzer
- Quantify libraries using qPCR for accurate sequencing concentration
- Include spike-in controls if quantifying absolute expression
Sequencing Quality Metrics:
- Monitor base quality scores throughout sequencing run
- Ensure >80% of bases have Q-score ≥30
- Check for adapter contamination and remove contaminated reads
Post-Sequencing QC:
- Use FastQC for initial quality assessment
- Employ appropriate trimming tools to remove low-quality bases and adapters
- Verify read alignment rates to reference genome
- Check for even coverage across transcripts
HCC-Specific QC Considerations
HCC tissues present unique challenges for quality control due to their complex microenvironment and heterogeneity. The following HCC-specific QC measures should be implemented:
Protocol: HCC-Specific Quality Control Measures
Tumor Purity Assessment:
- Perform histopathological review of adjacent tissue sections
- Document tumor cellularity percentage for each sample
- Consider digital pathology approaches for quantitative assessment
- Exclude samples with <60% tumor cellularity unless studying tumor microenvironment
Non-Hepatocyte Content Evaluation:
HCC Subtype Verification:
- Check expression of established HCC subtype markers
- Verify consistency with expected molecular profiles based on etiology
Inter-sample Contamination Check:
- Include genotype markers if available
- Use bioinformatic tools to detect sample mix-ups
The diagram below illustrates the comprehensive quality control workflow for HCC RNA sequencing studies:
Diagram 1: Comprehensive quality control workflow for HCC RNA sequencing studies, illustrating key decision points and quality thresholds.
Table 3: Key Research Reagent Solutions for HCC RNA Sequencing Studies
| Reagent/Technology | Specific Examples | Application in HCC Research |
|---|---|---|
| RNA Stabilization Reagents | RNAlater, PAXgene Tissue System | Preserve RNA integrity in HCC tissues during acquisition and storage [44] |
| Single-Cell Isolation Kits | 10x Genomics Chromium System, Takara ICELL8 | Enable single-cell transcriptomic profiling of HCC heterogeneity [44] [46] |
| RNA Extraction Kits | Qiagen RNeasy, Zymo Research Quick-RNA | High-quality RNA extraction from FFPE and frozen HCC tissues |
| Library Prep Kits | Illumina TruSeq, SMARTer Stranded Total RNA-Seq | Library preparation with ribosomal depletion for ncRNA capture |
| Immunohistochemistry Antibodies | HepPar1, Arg-1, GPC3, CD34 [47] | Validate hepatocellular differentiation and tumor characteristics [47] |
| Cell Line Models | Huh-7, HCCLM3, HCCLM9, HepG2 [44] [42] | In vitro functional validation of lncRNA candidates |
| Bioinformatics Tools | Seurat, CellChat, Monocle [44] [46] | Single-cell data analysis and cellular communication mapping [44] |
| Quality Control Instruments | Agilent Bioanalyzer, Qubit Fluorometer | Assess RNA integrity and quantify nucleic acid concentration |
Robust experimental design incorporating meticulous tissue acquisition protocols, appropriate sample size calculation, and comprehensive quality control procedures is fundamental to generating meaningful data in HCC RNA sequencing studies. The complex heterogeneity of HCC, with its varied etiologies and molecular subtypes, necessitates careful patient stratification and sample processing to ensure research findings are biologically relevant and reproducible. By implementing the standardized protocols and quality control frameworks outlined in this application note, researchers can enhance the reliability of their investigations into non-coding RNAs in HCC, ultimately contributing to improved understanding of HCC pathogenesis and the development of novel diagnostic and therapeutic strategies. As HCC research continues to evolve, particularly with advances in single-cell technologies and spatial transcriptomics, these foundational experimental design principles will remain essential for producing high-quality, clinically translatable research findings.
The emergence of advanced RNA sequencing technologies has revolutionized non-coding RNA (ncRNA) research in hepatocellular carcinoma (HCC). This application note provides a structured comparison between bulk and single-cell RNA sequencing (scRNA-seq) methodologies for ncRNA profiling, detailing their respective applications in discovering biomarkers, dissecting tumor heterogeneity, and understanding therapeutic resistance mechanisms. We present standardized protocols, analytical frameworks, and practical decision-making guidelines to assist researchers in selecting the optimal approach for their specific investigational needs in HCC research.
Hepatocellular carcinoma represents a formidable oncological challenge as the third leading cause of cancer-related deaths worldwide [48]. The complexity of HCC tumorigenesis involves substantial alterations in the non-coding transcriptome, including long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) that regulate gene expression through diverse mechanisms [49] [50]. These ncRNAs function as critical regulators of cellular processes, influencing chromatin organization, transcription, RNA processing, and signal transduction [49]. The comprehensive analysis of ncRNAs in HCC has been transformed by RNA sequencing technologies, which have evolved from bulk population-level analysis to high-resolution single-cell approaches, each offering distinct advantages for specific research applications [51].
Technology Comparison: Resolution, Applications, and Limitations
Fundamental Technical Differences
Bulk RNA-seq provides a population-average gene expression profile from a heterogeneous cell population, where RNA from multiple cell types is extracted, pooled, and sequenced simultaneously. This method yields a comprehensive overview of the transcriptome but cannot distinguish cell-to-cell expression variations [52] [53]. In contrast, single-cell RNA sequencing (scRNA-seq) isolates individual cells before RNA extraction and library preparation, enabling the resolution of gene expression at the individual cell level. The core technology of platforms like the 10X Genomics Chromium system involves generating hundreds of thousands of single-cell microdroplets (GEMs), each containing a single cell, reverse transcription mixes, and a gel bead conjugated with oligo sequences featuring cell-specific barcodes and unique molecular identifiers (UMIs) for precise transcript quantification [51].
Quantitative Comparison of Performance Characteristics
Table 1: Technical and Performance Comparison of Bulk vs. Single-Cell RNA-seq
| Feature | Bulk RNA-seq | Single-Cell RNA-seq |
|---|---|---|
| Resolution | Population average | Individual cell level |
| Cost per Sample | Lower (~$300/sample) | Higher (~$500-$2000/sample) [52] |
| Data Complexity | Lower | Higher, requiring specialized computational methods [52] |
| Cell Heterogeneity Detection | Limited | High [52] |
| Gene Detection Sensitivity | Higher (median ~13,378 genes/sample) | Lower (median ~3,361 genes/sample) [52] |
| Rare Cell Type Detection | Limited | Possible, even at frequencies of 1 in 10,000 cells [52] |
| Splicing Analysis | More comprehensive | Limited [52] |
| Sample Input Requirement | Higher | Lower, capable with picogram RNA quantities [52] |
| Theoretical Basis | Averages expression across all cells in sample | Deconvolutes expression using cell barcodes and UMIs |
Application-Specific Strengths and Limitations
Bulk RNA-seq demonstrates particular utility for:
- Differential gene expression analysis between conditions (e.g., tumor vs. normal)
- Transcriptome annotation and novel ncRNA discovery
- Alternative splicing and isoform analysis
- Fusion gene detection [52] [53]
- Large-scale biomarker studies with homogeneous samples
However, bulk approaches mask cellular heterogeneity and cannot identify rare cell populations, potentially obscuring crucial biological insights [52]. For example, bulk sequencing of glioblastoma samples failed to capture intratumoral heterogeneity that was subsequently revealed by scRNA-seq [52].
scRNA-seq excels in:
- Resolving cellular heterogeneity within complex tissues
- Identifying novel and rare cell types/states
- Constructing developmental trajectories and lineage tracing
- Characterizing tumor microenvironment composition
- Immune cell profiling and subset discovery [52] [53]
The limitations of scRNA-seq include higher costs, greater data complexity, technical challenges like dropout events, and lower gene detection sensitivity per cell [52].
Applications in HCC ncRNA Research: Evidence and Protocols
Bulk RNA-seq Applications and Protocols in HCC
Protocol 3.1.1: Genome-Wide lncRNA Profiling in HCC Tissues
Objective: Identify differentially expressed lncRNAs between HCC and paired normal tissues.
Experimental Workflow:
- Tissue Collection: Obtain fresh-frozen HCC and paired adjacent normal liver tissues (3 cm from tumor margin), confirmed by histological examination [54].
- RNA Extraction: Isolate total RNA using TRIzol reagent, ensuring RNA Integrity Number (RIN) >8.0.
- Library Preparation: Deplete ribosomal RNA and construct sequencing libraries using strand-specific protocols compatible with Illumina platforms.
- Sequencing: Perform 150bp paired-end sequencing on Illumina HiSeq2500, targeting ~45 million reads per sample [54].
- Bioinformatic Analysis:
- Align reads to reference genome (GRCh37) and lncRNA databases (NONCODE)
- Identify differentially expressed lncRNAs (fold change >2, FDR <0.05)
- Validate top candidates by qRT-PCR in independent cohort [54]
Key Findings: This approach identified 214 differentially expressed lncRNAs in HCC, including several (NONHSAT003823, NONHSAT056213, NONHSAT015386, NONHSAT122051) correlated with clinicopathological features like tumor differentiation, portal vein tumor thrombosis, and AFP levels [54].
Single-Cell RNA-seq Applications and Protocols in HCC
Protocol 3.2.1: scRNA-seq for Therapy Resistance-Associated ncRNAs
Objective: Identify ncRNAs mediating sorafenib resistance in HCC at single-cell resolution.
Experimental Workflow:
- Model Establishment: Generate sorafenib-resistant HCC cells (Hep3B-R, Huh7-R) through long-term drug exposure [55].
- Single-Cell Suspension: Prepare viable single-cell suspensions with >90% viability.
- Single-Cell Partitioning: Use 10X Genomics Chromium Controller for single-cell capture and barcoding.
- Library Preparation: Follow 10X Genomics protocol for GEM generation, reverse transcription, and cDNA amplification.
- Sequencing: Perform sequencing on Illumina platforms targeting 50,000 reads/cell.
- Bioinformatic Analysis:
- Process data using Cell Ranger pipeline
- Perform quality control, normalization, and clustering with Seurat
- Identify differentially expressed ncRNAs between sensitive and resistant cells [55]
Key Findings: This approach revealed that a small subpopulation of pre-existing quiescent stem-like cells with intrinsic sorafenib resistance expands under treatment pressure. The lncRNA ZFAS1 was identified as markedly upregulated in resistant cells and associated with stemness/EMT phenotypes and poor prognosis [55].
Protocol 3.2.2: Integrated scRNA-seq Analysis of HCC Ecosystem
Objective: Characterize the multicellular ecosystem of primary and metastatic HCC.
Experimental Workflow:
- Sample Processing: Process multiple tissue types (non-tumor liver, primary tumor, portal vein tumor thrombus, metastatic lymph node) from HCC patients [48].
- Cell Capture and Sequencing: Use 10X Genomics platform to capture ~2,000 cells/sample.
- Data Integration: Apply canonical correlation analysis (CCA) for batch correction across patients and tissues.
- Cell Type Annotation: Use shared-nearest neighbor clustering and marker gene identification.
- Tumor Microenvironment Analysis: Investigate cell-cell communication and spatial relationships.
Key Findings: This comprehensive atlas identified 14 distinct cell clusters, revealed enrichment of central memory T cells in early tertiary lymphoid structures associated with improved survival, and demonstrated distinct T-cell exhaustion patterns in HBV-related HCCs [48].
Integrated Approaches for Enhanced Resolution
Protocol 3.3.1: Combining scRNA-seq and Bulk RNA-seq for Biomarker Development
Objective: Develop prognostic biomarkers by integrating single-cell and bulk sequencing data.
Experimental Workflow:
- scRNA-seq Data Generation: Profile tumor and adjacent normal tissues to identify cell type-specific expression patterns.
- Cell Type Contribution Analysis: Calculate FCscores to quantify each cell subset's contribution to HCC development based on proportion and expression changes of signature genes [56].
- Bulk Validation: Validate findings in large bulk RNA-seq cohorts (TCGA-LIHC, GSE14520).
- Model Construction: Use LASSO Cox regression to develop prognostic signatures [56] [57].
Key Findings: Integration approaches have successfully identified plasma cells as key contributors to HCC development and facilitated the construction of prognostic models based on plasma cell-related genes [56].
Table 2: ncRNA Classes and Their Investigation Using RNA-seq Technologies in HCC
| ncRNA Class | Key Functions | Exemplary Findings in HCC | Optimal Method |
|---|---|---|---|
| lncRNAs | Chromatin modification, transcriptional regulation | PVT1 and SNHG7 promote HCC invasion [50]; ZFAS1 mediates sorafenib resistance [55] | Both (scRNA-seq for heterogeneity, bulk for discovery) |
| miRNAs | Post-transcriptional regulation, mRNA degradation | miR-142-3p reverses TKI resistance by targeting YES1/TWF1 [49] | Bulk RNA-seq with targeted validation |
| circRNAs | miRNA sponging, translation, protein scaffolding | circRNA-miRNA-mRNA networks influence HCC differentiation [49] | Bulk for comprehensive profiling, scRNA-seq for cellular specificity |
Table 3: Essential Research Reagents and Computational Tools for ncRNA Profiling
| Category | Specific Tool/Reagent | Function/Application | Example Use Cases |
|---|---|---|---|
| Wet Lab Reagents | 10X Genomics Chromium System | Single-cell partitioning and barcoding | Capturing transcriptomes of up to 20,000 individual cells [51] |
| Smart-seq2 Chemistry | Full-length cDNA generation from low RNA input | Sensitive detection of ncRNAs in rare cell populations [52] | |
| TRIzol/RNA Extraction Kits | High-quality RNA isolation | Preserving RNA integrity for accurate ncRNA quantification [54] | |
| Sequencing Platforms | Illumina HiSeq/NovaSeq | High-throughput sequencing | Bulk RNA-seq with 45M+ reads per sample [54] |
| 10X Genomics Chromium Controller | Single-cell library preparation | scRNA-seq of heterogeneous HCC tissues [51] | |
| Bioinformatic Tools | Seurat R Package | scRNA-seq data analysis | Dimensionality reduction, clustering, and differential expression [48] [56] |
| Cell Ranger | Processing 10X Genomics data | Initial processing of single-cell data [51] | |
| AUCell Algorithm | Calculating pathway activity scores | Assessing liquid-liquid phase separation activity in single cells [57] | |
| Monocle 2 | Trajectory inference | Reconstructing cellular differentiation paths [57] | |
| Databases | DrLLPS | Liquid-liquid phase separation genes | Identifying LLPS-related ncRNAs in HCC [57] |
| NONCODE | Reference lncRNA database | Annotating novel lncRNAs in HCC [54] | |
| TCGA-LIHC | HCC genomic data | Validation cohort for biomarker studies [56] |
Selection Guidelines
The choice between bulk and single-cell RNA-seq depends on research objectives, budget, sample characteristics, and analytical capabilities:
Choose Bulk RNA-seq when:
- Studying homogeneous cell populations or tissue-level expression patterns
- Budget constraints necessitate larger sample sizes
- Research focuses on differential expression between conditions
- Investigating alternative splicing, novel transcripts, or fusion genes
- Analytical resources for complex data processing are limited [52] [53]
Choose scRNA-seq when:
- Investigating cellular heterogeneity within complex tissues
- Identifying rare cell populations or states
- Tracing developmental trajectories or lineage relationships
- Characterizing tumor microenvironment composition
- Studying therapy resistance mechanisms and cancer stem cells [52] [53] [55]
Emerging Trends and Future Perspectives
The field of ncRNA profiling in HCC is rapidly evolving with several emerging trends:
- Multi-omics integration: Combining scRNA-seq with epigenetic and proteomic analyses
- Spatial transcriptomics: Mapping ncRNA expression within tissue architecture
- Liquid biopsy applications: Analyzing ncRNAs in circulating tumor cells and exosomes
- Therapeutic targeting: Developing ncRNA-based therapeutics for HCC treatment [49]
- Artificial intelligence: Implementing machine learning for pattern recognition in complex ncRNA data
As these technologies continue to advance and costs decrease, the integration of bulk and single-cell approaches will provide increasingly comprehensive insights into ncRNA biology in HCC, ultimately accelerating the development of novel diagnostic biomarkers and therapeutic strategies.
RNA sequencing (RNA-Seq) has become a foundational technology for probing the transcriptome, offering unparalleled insights into gene expression patterns. Its application in oncology, particularly in the study of non-coding RNAs (ncRNAs) in hepatocellular carcinoma (HCC), is driving the discovery of novel diagnostic biomarkers and therapeutic targets [2]. Hepatocellular carcinoma is a malignancy with increasing global incidence and mortality, characterized by a complex molecular landscape where dysregulated long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) play crucial oncogenic or tumor-suppressive roles [2] [8]. The analysis of these molecules from tissue samples requires a robust and reproducible bioinformatics workflow. This protocol provides a detailed, step-by-step guide for a computational pipeline that processes raw RNA-Seq data from HCC tissues, transforming it into biologically meaningful information about differential ncRNA expression, thereby empowering research into liver cancer pathogenesis.
The bioinformatics pipeline for RNA-Seq analysis is a multi-stage process. It begins with raw sequencing data and culminates in a list of confidently identified differentially expressed genes (DEGs). The entire workflow can be conceptually divided into two major phases: initial data processing and differential expression analysis, each involving critical quality control checkpoints.
The following diagram illustrates the complete workflow, from raw sequencing files to final visualization, highlighting the key stages and software used at each step.
Phase 1: Data Preparation – From FASTQ to Count Matrix
The initial phase is computationally intensive and is typically performed in a Unix-like command-line environment, often on a high-performance computing cluster [58] [59]. This phase focuses on ensuring data quality and generating accurate gene-level counts.
Software Installation and Setup
Begin by installing the necessary bioinformatics tools. Using a package manager like Bioconda significantly simplifies this process and ensures dependency resolution [58].
Quality Control and Read Trimming
The first analytical step is to assess the quality of the raw sequencing data contained in FASTQ files. FastQC is the standard tool for this, generating a comprehensive HTML report on read quality, adapter contamination, and other potential issues [58]. Following quality assessment, Trimmomatic is used to remove low-quality bases, adapter sequences, and other artifacts, which improves the reliability of downstream alignment [58].
Alignment to a Reference Genome
The trimmed and cleaned sequencing reads must be aligned to a reference genome. For RNA-Seq data, a splice-aware aligner is essential to accurately map reads across exon-intron boundaries. HISAT2 is a widely used, memory-efficient aligner for this purpose [58]. Alternatively, STAR is another powerful and highly accurate aligner, though it requires more memory [59]. The result of this step is a Sequence Alignment Map (SAM) file, which is then converted to its binary counterpart (BAM) and sorted using samtools [58].
Quantification of Gene Abundance
After alignment, the number of reads mapping to each gene is counted. featureCounts (part of the Subread package) is a common tool that uses a genome annotation file (GTF/GFF) to assign aligned reads to genomic features, such as genes or non-coding RNA loci, producing a raw count table [58]. For a more nuanced analysis that accounts for transcript-level ambiguity, alignment-free quantifiers like Salmon can be used, which often leads to improved accuracy and speed [59].
Phase 2: Differential Expression Analysis in R
The count matrix generated in Phase 1 serves as the input for statistical analysis in R. This phase identifies genes, including ncRNAs, whose expression is significantly altered between conditions (e.g., HCC tumor vs. non-tumor liver tissue).
Loading and Preparing Data
The first step in R is to load the count data and associated sample information (metadata). The metadata should describe the experimental conditions for each sample. It is critical to check that the sample names in the count matrix and metadata are consistent.
Statistical Testing with DESeq2
The DESeq2 package is a standard tool for differential expression analysis from count data. It uses a negative binomial generalized linear model to estimate variance and test for significance, while internally correcting for library size differences [60]. The analysis involves creating a DESeqDataSet object, running the DESeq2 pipeline, and extracting the results.
Visualization of Results
Effective visualization is key to interpreting the results of a differential expression analysis. Common plots include the volcano plot, which displays the relationship between statistical significance (-log10 p-value) and magnitude of change (log2 fold change), and the heatmap, which shows expression patterns of top DEGs across all samples, revealing sample clustering [58] [60].
The Scientist's Toolkit: Essential Research Reagents and Software
The following table details key software, reagents, and resources required to execute the RNA-Seq data analysis pipeline for HCC ncRNA research.
Table 1: Essential Reagents and Software for RNA-Seq Analysis of HCC
| Item Name | Function/Description | Application Note |
|---|---|---|
| FastQC [58] | Quality control tool for high-throughput sequence data. | Assesses raw sequence data from FASTQ files; critical for identifying sequencing errors or adapter contamination before proceeding. |
| Trimmomatic [58] | Flexible read trimming tool for Illumina NGS data. | Removes adapter sequences and low-quality bases to improve the quality of downstream alignment. |
| HISAT2 [58] | Fast and sensitive splice-aware aligner for NGS data. | Aligns RNA-Seq reads to a reference genome (e.g., GRCh38), accounting for introns. A key alternative is STAR [59]. |
| featureCounts [58] | Efficient program for assigning sequence reads to genomic features. | Generates the count matrix by summarizing reads mapped to genes or ncRNA loci defined in a GTF annotation file. |
| R/Bioconductor [60] | Programming environment for statistical computing and genomics. | The primary platform for differential expression analysis and visualization (e.g., with DESeq2, limma, ggplot2). |
| DESeq2 [60] | R package for differential analysis of count data. | Uses a negative binomial model to determine statistically significant differentially expressed genes/ncRNAs between conditions. |
| Reference Genome & Annotation | Species-specific genomic sequence and gene model file (GTF/GFF). | Provides the coordinate system for alignment (FASTA) and defines gene/transcript features for quantification. Essential for ncRNA analysis. |
| nf-core/rnaseq [59] | A community-built, portable pipeline for RNA-Seq data analysis. | Automates the entire data preparation phase (QC, alignment, quantification), ensuring reproducibility and best practices. |
Interpreting Results in the Context of HCC ncRNA Biology
The final output of this pipeline is a ranked list of differentially expressed ncRNAs. In HCC research, this list must be interpreted through the lens of existing biological knowledge. For instance, the pipeline might identify the downregulation of the tumor suppressor miR-122 or the upregulation of the oncogenic LINC00152 [2] [8]. The high diagnostic accuracy (100% sensitivity, 97% specificity) achieved by machine learning models integrating lncRNA expression profiles underscores the potential clinical utility of such findings [8].
The analytical workflow for interpreting differential expression results involves validating key findings and placing them within established biological pathways, as illustrated below.
Subsequent functional enrichment analysis (e.g., Gene Ontology, KEGG pathways) can reveal if the dysregulated ncRNAs are associated with HCC-relevant processes such as cell proliferation, metastasis, or known oncogenic signaling pathways like AKT and VEGF, which are commonly targeted in HCC therapeutics [2] [61]. This integrated, step-by-step pipeline provides a solid foundation for unlocking the molecular secrets of hepatocellular carcinoma through the lens of non-coding RNA biology.
Integrating scRNA-seq and Bulk RNA-seq to Deconvolute HCC Heterogeneity
Hepatocellular carcinoma (HCC) represents a paradigm of complex tumor heterogeneity, characterized by diverse cellular subpopulations and dynamic tumor microenvironment (TME) interactions that drive progression and therapeutic resistance [48]. The integration of single-cell RNA sequencing (scRNA-seq) and bulk RNA sequencing (bulk RNA-seq) has emerged as a powerful methodological framework to dissect this complexity, bridging high-resolution cellular characterization with population-level clinical correlations [57] [62]. This approach is particularly valuable for investigating non-coding RNAs in HCC tissues, as it enables researchers to pinpoint their cell-type-specific expression patterns and functional roles within the broader pathological context.
The fundamental challenge in HCC research lies in its substantial cellular diversity. Bulk RNA-seq provides comprehensive transcriptomic data but averages expression profiles across all cells, potentially obscuring critical cell-type-specific signals [63]. Conversely, scRNA-seq resolves cellular heterogeneity at unprecedented resolution but often lacks the statistical power for robust clinical association studies [62]. Integrative methodologies overcome these limitations by leveraging the strengths of both approaches, creating a more complete picture of HCC biology that informs both biomarker discovery and therapeutic development.
Key Analytical Frameworks and Methodologies
Data Processing and Quality Control Pipeline
The initial phase of integrative analysis requires rigorous processing of scRNA-seq data to ensure data quality and reliability. The standard workflow utilizes the Seurat package (versions 4.0-5.1.0) for quality control, normalization, and batch correction [46] [57] [64]. Critical quality control parameters typically include:
- Cell Filtering: Exclusion of cells with <200 or >4000-6000 detected genes and mitochondrial gene content >10-15% [46] [62] [64]
- Gene Filtering: Removal of genes expressed in fewer than 3 cells [62]
- Normalization: Data normalization using the
NormalizeDatafunction or SCTransform method [57] [64] - Batch Correction: Application of Harmony algorithm or CCA (Canonical Correlation Analysis) for integrating multiple samples [63] [65]
Following quality control, dimensionality reduction is performed using Principal Component Analysis (PCA), typically retaining the first 10-30 principal components based on elbow plot inspection [46] [66]. Cell clustering is then conducted using graph-based methods such as the Louvain algorithm with resolution parameters ranging from 0.5 to 0.8 [62] [64]. Cell type annotation is performed by referencing canonical marker genes and databases like CellMarker 2.0 and Cell Taxonomy [64].
Integrative Computational Approaches
Several computational strategies have been successfully implemented to integrate scRNA-seq and bulk RNA-seq data in HCC research:
- Cell Type Deconvolution: CIBERSORT and CIBERSORTx algorithms leverage scRNA-seq-derived cell signatures to estimate cell type abundances from bulk RNA-seq data, enabling correlation of specific cell populations with clinical outcomes [48] [67].
- Consensus Clustering: Molecular subtypes are identified through consensus clustering of bulk RNA-seq data based on gene sets derived from scRNA-seq analyses, such as lipid metabolism-related genes or senescence-associated genes [46] [65].
- Risk Model Construction: Machine learning approaches, particularly LASSO Cox regression, are applied to bulk RNA-seq data to develop prognostic signatures using genes identified from scRNA-seq studies [57] [62] [63].
- Cellular Communication Analysis: The CellChat package quantifies ligand-receptor interactions across cell types using scRNA-seq data, revealing how specific cell populations influence the TME [46] [57] [64].
Table 1: Key Computational Tools for Integrative Analysis
| Tool/Package | Primary Function | Application in HCC Research |
|---|---|---|
| Seurat | scRNA-seq processing and analysis | Cell type identification, dimensionality reduction [46] |
| CellChat | Cell-cell communication inference | Ligand-receptor interaction analysis [57] |
| CIBERSORT | Cell type deconvolution | Immune infiltration estimation from bulk data [48] |
| Monocle | Trajectory inference | Pseudotemporal ordering of cell states [57] |
| MOVICS | Multi-omics integration | HCC subtyping using multiple algorithms [67] |
Experimental Protocols and Workflows
Protocol 1: scRNA-seq Data Processing and Cell Type Annotation
Purpose: To process raw scRNA-seq data from HCC tissues and identify distinct cell populations.
Materials:
- Raw scRNA-seq data (FASTQ files)
- Computing infrastructure with sufficient RAM (≥32GB recommended)
- R environment (v4.3.1 or higher) with Seurat package (v5.0.1)
Procedure:
- Data Import: Load gene-cell count matrices using
Read10X()function or create Seurat object directly withCreateSeuratObject()[62] [65]. - Quality Control: Filter cells based on the following criteria:
- Normalization: Normalize data using
NormalizeData()with normalization method "LogNormalize" and scale factor 10,000. - Feature Selection: Identify 2,000-4,000 highly variable genes using
FindVariableFeatures()with selection method "vst" [57] [65]. - Dimensionality Reduction:
- Run PCA using
RunPCA()with npcs=50 - Select principal components based on elbow plot visualization
- Perform UMAP/t-SNE using
RunUMAP()orRunTSNE()with appropriate dimensions [62]
- Run PCA using
- Clustering:
- Construct shared nearest neighbor graph with
FindNeighbors()using selected PCs - Identify clusters with
FindClusters()at resolution 0.5-0.8 [63]
- Construct shared nearest neighbor graph with
- Cell Type Annotation:
- Identify cluster-specific marker genes using
FindAllMarkers()(Wilcoxon test, min.pct=0.25, logfc.threshold=0.5) - Annotate cell types based on canonical marker expression [48]
- Identify cluster-specific marker genes using
Troubleshooting Tips:
- High mitochondrial percentage may indicate stressed/dying cells
- Low number of detected genes per cell may indicate empty droplets
- Batch effects can be corrected using Harmony or CCA integration [63]
Protocol 2: Integration with Bulk RNA-seq for Prognostic Model Development
Purpose: To develop a prognostic gene signature for HCC by integrating scRNA-seq and bulk RNA-seq data.
Materials:
- Processed scRNA-seq data with cell type annotations
- Bulk RNA-seq data with clinical outcomes (e.g., TCGA-LIHC, ICGC)
- R packages: limma, survival, glmnet, clusterProfiler
Procedure:
- Identify Cell-Type-Specific Genes:
- Bulk RNA-seq Differential Expression:
- Gene Set Intersection:
- Intersect cell-type-specific genes from scRNA-seq with bulk DEGs
- Perform survival analysis on intersected genes using Cox proportional hazards model [63]
- Prognostic Model Construction:
- Model Validation:
- Functional Enrichment:
Expected Outcomes:
- A multivariate gene signature predictive of HCC patient survival
- Identification of key biological processes associated with high-risk patients
- Potential therapeutic targets for further validation
Key Signaling Pathways and Cellular Interactions
PTGES3-Mediated Immunosuppressive Signaling
Recent integrative analyses have identified PTGES3 as a central regulator in lipid metabolism-reprogrammed HCC cells, where it facilitates immunosuppression through specific ligand-receptor interactions [46]. The signaling mechanism can be summarized as follows:
Diagram 1: PTGES3 immunosuppressive signaling in HCC. PTGES3 enhances FN1 and MDK expression, which bind to CD44 and NCL receptors respectively on T cells, promoting immunosuppression [46].
This pathway demonstrates how malignant hepatocytes with altered lipid metabolism can manipulate the TME through PTGES3-mediated signaling, ultimately leading to T cell dysfunction and immunotherapy resistance [46].
APOA1-TREM2 and VTN-PLAUR in Hepatocyte-Macrophage Crosstalk
Multi-omics approaches have revealed critical communication axes between different cellular compartments in HCC. Two particularly significant ligand-receptor pairs are:
Diagram 2: Intercellular crosstalk in HCC TME. Hepatocytes and cholangiocytes communicate with macrophages via APOA1-TREM2 and VTN-PLAUR interactions, promoting pro-tumorigenic macrophage functions [67].
These interactions represent potential therapeutic targets for disrupting protumorigenic communication within the HCC ecosystem.
Data Presentation and Quantitative Findings
Experimentally Validated Prognostic Signatures
Integrative analyses have yielded multiple prognostic signatures with clinical validation. The table below summarizes key gene signatures derived from integrated scRNA-seq and bulk RNA-seq approaches:
Table 2: Experimentally Validated Prognostic Signatures in HCC
| Study Focus | Genes in Signature | Validation Approach | Clinical Utility |
|---|---|---|---|
| Lipid Metabolism [46] | PTGES3 (and 17 others) | Molecular docking; in vitro functional assays | Prognostic biomarker and therapeutic target |
| T Cell-Related [62] | PTTG1, LMNB1, SLC38A1, BATF | IHC in 25 patient tissues; external validation | Stratifies patients into high/low risk groups |
| RFS Prediction [63] | CDKN2A, CFHR3, CYP2C9, HMGÎ’2, IGLC2, JPT1 | RT-qPCR; independent cohort validation | Predicts recurrence-free survival |
| Cellular Senescence [65] | PTTG1 (and 3 others) | Immune infiltration analysis; functional assays | Links senescence to immune evasion |
| Macrophage-Associated [64] | KLK11, MARCO, CFP, KRT19, GAS1, SOD3, CYP2C8, TOP2A, CENPF, MKI67, NUPR1 | Machine learning models (Lasso, RF, XGBoost) | Early diagnosis from cirrhosis to HCC |
Cell Type Composition in HCC Ecosystem
scRNA-seq studies have quantitatively characterized the cellular heterogeneity of HCC. The following table summarizes key immune and stromal populations identified across multiple studies:
Table 3: Cellular Composition of HCC Tumor Microenvironment
| Cell Type | Subpopulations Identified | Key Markers | Functional Significance |
|---|---|---|---|
| T/NK Cells [48] | CTLs, MAIT, TEM, TRM, Treg, TCM | CD3D, CD8A, CD4, FOXP3, CCR7 | TCM enriched in early tertiary lymphoid structures; Tregs immunosuppressive |
| Myeloid Cells [48] | MMP9+ TAMs, MoMs, KCs | CD68, MMP9, PPARG | PPARγ drives TAM differentiation; MMP9+ TAMs terminally differentiated |
| Malignant Hepatocytes [57] | High-LLPS, Low-LLPS | AFP, GPC3, EPCAM, SPP1 | LLPS score associated with malignant differentiation |
| B Cells [48] | CD20+ B cells | MS4A1, CD79A | Co-localize with TCM in tertiary lymphoid structures |
| Fibroblasts [66] | CAFs | ACTA2, FAP, PDGFRB | Contribute to extracellular matrix remodeling |
The Scientist's Toolkit
Essential Research Reagent Solutions
Table 4: Key Reagents and Computational Resources for HCC Integration Studies
| Resource | Type | Function | Specific Application |
|---|---|---|---|
| Seurat Suite [46] | R Package | scRNA-seq analysis | Quality control, normalization, clustering, differential expression |
| CellChatDB [57] | Database | Ligand-receptor interactions | Inference of cell-cell communication networks |
| CellAge [65] | Database | Senescence-associated genes | Identification of senescence-related signatures in HCC |
| DrLLPS [57] | Database | Liquid-liquid phase separation genes | Calculation of LLPS scores in malignant hepatocytes |
| TCGA-LIHC [46] | Data Resource | Bulk RNA-seq with clinical data | Model training and validation (n=370 HCC, 50 normal) |
| GEO GSE149614 [46] | Data Resource | scRNA-seq from 10 HCC patients | TME ecosystem mapping across multiple tissue sites |
| CIBERSORTx [48] | Algorithm | Digital cell fractionation | Deconvolution of bulk RNA-seq using scRNA-seq signatures |
| MOVICS [67] | R Package | Multi-omics integration | HCC subtyping using ten consensus clustering algorithms |
The integration of scRNA-seq and bulk RNA-seq technologies provides an powerful framework for deconvoluting HCC heterogeneity, revealing critical insights into cell-type-specific regulatory networks, TME dynamics, and molecular drivers of disease progression. The methodologies and findings summarized in this application note demonstrate how this integrated approach can identify clinically actionable biomarkers and therapeutic targets, ultimately advancing precision oncology for HCC patients. As these technologies continue to evolve, they will undoubtedly yield deeper understanding of non-coding RNA functions in specific cellular contexts within HCC tissues, opening new avenues for therapeutic intervention.
Leveraging Machine Learning for Diagnostic and Prognostic Biomarker Identification
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and a leading cause of cancer-related deaths worldwide [68] [69]. A significant challenge in managing HCC is the frequent diagnosis at advanced stages, where curative treatment options are limited. This is largely because current standard diagnostic methods, including ultrasound imaging and serum alpha-fetoprotein (AFP) measurement, lack sufficient sensitivity and specificity for early detection [70] [69]. There is an urgent need for more accurate diagnostic and prognostic biomarkers to enable earlier intervention and improve patient outcomes.
The emergence of high-throughput transcriptomic technologies, particularly RNA sequencing (RNA-Seq), has provided unprecedented opportunities for biomarker discovery. These technologies can generate comprehensive profiles of coding and non-coding RNAs in tissues and biofluids [70] [71]. Machine learning (ML) has become an indispensable tool for analyzing these complex, high-dimensional datasets to identify subtle but biologically significant patterns associated with disease states [68]. By applying sophisticated computational algorithms to RNA-Seq data, researchers can now identify robust biomarker signatures that outperform traditional markers like AFP, paving the way for more precise HCC management [71] [72].
Key Diagnostic Biomarkers Identified Through Machine Learning
Machine learning approaches have successfully identified numerous molecular biomarkers for HCC diagnosis, spanning protein-coding genes, non-coding RNAs, and multi-analyte signatures. The application of ML techniques such as support vector machine recursive feature elimination (SVM-RFE) and random forest with recursive feature elimination (RF-RFE) to transcriptomic data has significantly enhanced the identification of diagnostically relevant features.
Table 1: Diagnostic Protein-Coding Gene Biomarkers for HCC Identified by Machine Learning
| Gene Symbol | Full Name | AUC Range | Biological Function | Selection Method |
|---|---|---|---|---|
| CDKN3 | Cyclin Dependent Kinase Inhibitor 3 | >0.81 | Cell cycle regulation | SVM-RFE, RF-RFE [73] [74] |
| TRIP13 | Thyroid Hormone Receptor Interactor 13 | >0.81 | Mitotic regulation | SVM-RFE, RF-RFE [73] [74] |
| RACGAP1 | Rac GTPase Activating Protein 1 | >0.81 | Cytokinesis regulation | SVM-RFE, RF-RFE [73] [74] |
| SLC6A8 | Solute Carrier Family 6 Member 8 | Not specified | Creatine transport | LASSO, SVM-RFE, RF-Boruta [75] |
| PARP2-202 | Poly(ADP-Ribose) Polymerase 2 Transcript | Not specified | DNA repair | RF, SVM-RFE [71] |
| SPON2-203 | Spondin 2 Transcript | Not specified | Extracellular matrix protein | RF, SVM-RFE [71] |
Table 2: Diagnostic Non-Coding RNA Biomarkers for HCC
| ncRNA Category | Specific Biomarkers | Sample Source | Performance (AUC) | Reference |
|---|---|---|---|---|
| MicroRNAs (miRNAs) | miR-21, miR-224, miR-122, miR-9-3p | Plasma, Serum | 0.773-0.96 [70] | |
| Long Non-coding RNAs (lncRNAs) | LINC00152, UCA1, LINC00853, GAS5 | Plasma | Individual: Moderate; Combined with ML: 1.0 [8] | |
| Circular RNAs (circRNAs) | Potential candidates identified | Body fluids | Under investigation [70] |
The integration of these molecular markers with standard clinical parameters through machine learning models has demonstrated remarkable diagnostic performance. For instance, a random forest model incorporating just seven clinical predictors (age, albumin, alkaline phosphatase, AFP, DCP, AST, and platelet count) achieved an accuracy of 98.9% and an AUC of 0.99 in detecting HCC [72]. Similarly, combining lncRNA expression profiles with conventional laboratory data using a machine learning framework resulted in 100% sensitivity and 97% specificity for HCC diagnosis [8].
Prognostic Biomarkers and Gene Signatures
Beyond diagnosis, machine learning has enabled the development of robust prognostic signatures that predict clinical outcomes for HCC patients. These biomarkers help stratify patients based on their risk of disease progression, recurrence, or mortality, facilitating personalized treatment approaches.
Table 3: Prognostic Gene Signatures in HCC Identified by Machine Learning
| Gene Signature | Component Genes | Prognostic Value | Selection Method |
|---|---|---|---|
| MCC Prognostic Signature | BCAT1, DPF1, CDKN2B, CDKN2C, TUBA3C, IGF1, CDC14B, SMARCA2 | Predicts overall survival; independent of AJCC stage | Univariate Cox + LASSO Cox [73] [74] |
| Single-Gene Prognostic Markers | APOE, ALB (favorable); XIST, FTL (unfavorable) | Overall survival prediction | scRNA-seq + Survival Analysis [76] |
| lncRNA Ratio Marker | LINC00152 to GAS5 expression ratio | Higher ratio correlated with increased mortality | qRT-PCR + Clinical correlation [8] |
The mitotic cell cycle (MCC) prognostic signature exemplifies the power of ML-driven biomarker discovery. This eight-gene signature was developed through univariate Cox regression followed by LASSO Cox regression analysis and validated in independent cohorts. The risk score derived from this signature proved to be an independent prognostic factor, and its combination with AJCC stage further improved prognostic accuracy [73] [74]. Kaplan-Meier analysis confirmed that high-risk scores were consistently associated with poorer survival across various clinical subgroups, including different stages, grades, ages, and genders [74].
Single-cell RNA sequencing combined with artificial intelligence has further refined prognostic assessment by identifying cell-type-specific expression patterns associated with clinical outcomes. Genes such as APOE and ALB are linked to better prognosis, while XIST and FTL expression correlate with poor survival [76]. This single-cell resolution provides unprecedented insights into the tumor microenvironment and its influence on disease progression.
Experimental Protocols for Biomarker Discovery and Validation
Sample Preparation and RNA Sequencing
Protocol: Tissue Collection and RNA Extraction
- Sample Collection: Obtain HCC tissues and matched adjacent normal tissues from patients undergoing surgical resection. Immediately snap-freeze tissues in liquid nitrogen and store at -80°C until RNA extraction [75].
- RNA Extraction: Use miRNeasy Mini Kit or equivalent for total RNA isolation. Include DNase treatment to remove genomic DNA contamination.
- Quality Control: Assess RNA integrity using Bioanalyzer or TapeStation. Accept only samples with RNA Integrity Number (RIN) >7.0 for sequencing.
- Library Preparation: Construct sequencing libraries using TruSeq Stranded Total RNA Library Prep Kit or equivalent. For coding RNA analysis, use ribodepletion to preserve non-coding RNA species.
- Sequencing: Perform paired-end sequencing (2×150 bp) on Illumina platform to a minimum depth of 30 million reads per sample.
Protocol: Liquid Biopsy for Circulating ncRNAs
- Blood Collection: Draw peripheral blood into EDTA tubes. Process within 2 hours of collection.
- Plasma/Serum Separation: Centrifuge at 1,900×g for 10 minutes at 4°C. Transfer supernatant to fresh tubes and recentrifuge at 16,000×g for 10 minutes to remove residual cells.
- RNA Isolation: Extract circulating RNAs using miRNeasy Serum/Plasma Kit or equivalent. Add carrier RNA during isolation to improve yield.
- Quality Assessment: Verify RNA quality using capillary electrophoresis. Circulating RNAs typically show degraded profiles, but specific ncRNAs remain stable.
- Library Preparation: Use specialized kits optimized for low-input RNA (e.g., SMARTer smRNA-Seq Kit) to capture ncRNA species.
Machine Learning Analysis Pipeline
Protocol: Feature Selection using SVM-RFE and RF-RFE
- Data Preprocessing:
SVM-RFE Implementation:
RF-RFE Implementation:
Model Validation:
Protocol: Prognostic Model Development using LASSO Cox Regression
- Survival Data Preparation:
- Include patients with follow-up time ≥30 days.
- Calculate overall survival from diagnosis date to death or last follow-up.
Feature Pre-selection:
- Perform univariate Cox regression on all genes.
- Exclude genes violating proportional hazards assumption (Schoenfeld residuals p≤0.05).
- Apply Benjamini-Hochberg FDR correction; retain genes with FDR<0.05.
LASSO Cox Regression:
Risk Score Calculation:
Validation Using Quantitative Methods
Protocol: qRT-PCR Validation for Candidate Biomarkers
- cDNA Synthesis:
- Use 500ng-1μg total RNA for reverse transcription with RevertAid First Strand cDNA Synthesis Kit.
- Apply random hexamers and oligo-dT primers for comprehensive cDNA representation.
qPCR Reaction:
- Use PowerTrack SYBR Green Master Mix on ViiA 7 Real-Time PCR System.
- Perform reactions in triplicate with the following cycling conditions: 95°C for 2min, followed by 40 cycles of 95°C for 15sec and 60°C for 1min.
Primer Design:
- Design primers spanning exon-exon junctions to minimize genomic DNA amplification.
- Validate primer specificity using melt curve analysis and gel electrophoresis.
Data Analysis:
- Calculate relative expression using the 2^(-ΔΔCt) method with GAPDH or RN18S as reference genes.
- Perform ROC analysis to determine diagnostic accuracy of individual markers.
Visualization of Experimental Workflows and Signaling Pathways
ML-Driven Biomarker Discovery Workflow
Key Signaling Pathways in HCC Biomarkers
Table 4: Essential Research Reagents and Computational Tools for HCC Biomarker Discovery
| Category | Specific Product/Platform | Application | Key Features |
|---|---|---|---|
| RNA Isolation Kits | miRNeasy Mini Kit (QIAGEN) | Total RNA extraction from tissues | Preserves miRNA fraction |
| miRNeasy Serum/Plasma Kit (QIAGEN) | Circulating RNA isolation | Optimized for low-concentration samples | |
| Library Prep Kits | TruSeq Stranded Total RNA Kit (Illumina) | RNA-seq library construction | Ribodepletion for ncRNA analysis |
| SMARTer smRNA-Seq Kit (Takara Bio) | Small RNA sequencing | Specifically captures miRNAs | |
| qRT-PCR Reagents | PowerTrack SYBR Green Master Mix (Applied Biosystems) | Gene expression validation | Sensitive detection with wide dynamic range |
| TaqMan MicroRNA Assays (Thermo Fisher) | miRNA quantification | Specific detection of mature miRNAs | |
| Computational Tools | TCGAbiolinks (R/Bioconductor) | TCGA data access and analysis | Streamlined interface to NCI genomic data |
| edgeR, limma (R/Bioconductor) | Differential expression analysis | Robust statistical methods for RNA-seq | |
| caret, e1071 (R/CRAN) | Machine learning implementation | Unified interface for multiple ML algorithms | |
| ImmuCellAI (R/Python) | Immune cell infiltration analysis | Deconvolution of 24 immune cell types |
Machine learning has revolutionized the identification of diagnostic and prognostic biomarkers for hepatocellular carcinoma, enabling the discovery of molecular signatures with superior performance compared to conventional markers. The integration of transcriptomic data from both tissue and liquid biopsies with sophisticated computational algorithms has yielded robust biomarker panels that can accurately detect HCC and predict patient outcomes.
Future directions in this field will likely focus on several key areas: (1) the integration of multi-omics data (genomics, transcriptomics, proteomics) to develop more comprehensive biomarker signatures; (2) the application of deep learning and artificial intelligence to single-cell RNA sequencing data to decipher cellular heterogeneity in the tumor microenvironment; (3) the development of standardized protocols for liquid biopsy-based biomarkers to enable non-invasive monitoring of treatment response and disease recurrence; and (4) the implementation of these biomarkers in clinical trials to validate their utility in guiding personalized treatment decisions. As these technologies continue to evolve, ML-driven biomarker discovery will play an increasingly central role in improving early detection and personalized management of HCC.
Overcoming Analytical Hurdles in HCC ncRNA Data Interpretation
The reliability of RNA sequencing (RNA-seq) data is often undermined by batch effects—systematic non-biological differences that arise during sample processing and sequencing across different batches [77]. These technical variations can be on a similar scale or even larger than the biological differences of interest, significantly reducing the statistical power to detect genuinely differentially expressed (DE) genes [77]. In the context of hepatocellular carcinoma (HCC) research, where investigators frequently analyze non-coding RNAs (ncRNAs) from clinical tissues, batch effects pose a substantial challenge. Specimens collected over extended periods, processed by different personnel, or sequenced in multiple runs can introduce technical noise that obscures the subtle expression patterns of ncRNAs, which are crucial regulators in HCC pathogenesis [2] [36].
Addressing batch effects is not merely an optional refinement but a critical necessity for ensuring data integrity. Batch effects can stem from various sources, including different library preparation kits, sequencing platforms, reagent lots, personnel, or time of processing [78] [79]. For ncRNA-focused studies in HCC, this is particularly pertinent, as the accurate quantification of molecules like long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) is essential for identifying bona fide biomarkers and therapeutic targets [8] [2]. This Application Note provides a structured overview of batch effect correction and normalization strategies, framing them within established RNA-seq workflows to enhance the accuracy and interpretability of HCC transcriptomic data.
Key Concepts: Normalization vs. Batch Effect Correction
It is crucial to distinguish between normalization and batch effect correction, as they address distinct types of technical variations within RNA-seq data.
Normalization primarily adjusts for differences in sequencing depth and library composition between samples. The raw counts in a gene expression matrix cannot be directly compared because the number of reads mapped to a gene depends not only on its expression level but also on the total number of sequencing reads obtained for that sample [80]. Normalization techniques mathematically adjust these counts to remove such biases.
Table 1: Common Normalization Methods in RNA-seq Analysis
| Method | Sequencing Depth Correction | Gene Length Correction | Library Composition Correction | Suitable for DE Analysis | Key Characteristics |
|---|---|---|---|---|---|
| CPM | Yes | No | No | No | Simple scaling by total reads; affected by highly expressed genes |
| RPKM/FPKM | Yes | Yes | No | No | Adjusts for gene length; still affected by library composition |
| TPM | Yes | Yes | Partial | No | Scales sample to constant total; good for cross-sample comparison |
| Median-of-Ratios (DESeq2) | Yes | No | Yes | Yes | Robust to composition biases; uses a pseudo-reference sample |
| TMM (edgeR) | Yes | No | Yes | Yes | Trims extreme genes; robust to imbalances in highly differential expression |
In contrast, batch effect correction is a subsequent step that addresses systematic variations between groups of samples processed or sequenced in different batches. As noted in the Griffith Lab protocol, "batch effects in composition, i.e., the level of expression of genes scaled by the total expression (coverage) in each sample, cannot be fully corrected with normalization" [78]. Therefore, even after normalization, individual genes may still be affected by batch-level biases that require specific statistical correction methods [78].
Batch Effect Correction Strategies and Tools
Algorithm-Based Correction Methods
Several computational tools have been developed specifically to model and remove batch effects from RNA-seq count data while preserving biological signals.
The ComBat family of algorithms is widely used for this purpose. The original ComBat method employs an empirical Bayes framework to correct for both additive and multiplicative batch effects [77]. ComBat-seq extends this approach by using a negative binomial generalized linear model (GLM), which is more appropriate for RNA-seq count data, and has the advantage of preserving the integer nature of the count matrix, making it suitable for downstream DE analysis with tools like edgeR and DESeq2 [77] [78].
A recently developed refinement, ComBat-ref, builds upon ComBat-seq but introduces a key innovation: it selects a reference batch with the smallest dispersion and preserves the count data for this batch while adjusting other batches towards this reference [77]. This strategy demonstrates superior performance in both simulated environments and real-world datasets, significantly improving sensitivity and specificity compared to existing methods [77]. The method's effectiveness is attributed to its accurate modeling of count data using negative binomial distributions and its strategic use of a low-dispersion reference batch.
Machine learning-based approaches offer an alternative strategy. One method leverages a quality-aware approach by using a machine learning classifier (seqQscorer) to predict sample quality (Plow scores) and then uses these quality scores to detect and correct batch effects [79]. This quality-based correction was found to be comparable or superior to traditional batch correction in 92% of the tested datasets, particularly when coupled with outlier removal [79]. This approach is valuable when detailed batch information is unavailable, as it can detect batches based on quality differences between samples.
Table 2: Comparison of Batch Effect Correction Methods for RNA-seq Data
| Method | Statistical Foundation | Preserves Count Data | Key Advantage | Considerations |
|---|---|---|---|---|
| ComBat | Empirical Bayes (linear model) | No | Established method; handles additive/multiplicative effects | Designed for normalized data; not for raw counts |
| ComBat-seq | Negative Binomial GLM | Yes | Preserves integer counts; suitable for DE analysis | Performance can drop with high batch dispersion variance |
| ComBat-ref | Negative Binomial GLM with reference batch | Yes | Superior power with high-dispersion batches; preserves reference batch | Requires a low-dispersion reference batch for optimal performance |
| Quality-Aware ML | Machine learning-based quality prediction | Depends on implementation | Does not require prior batch information; uses quality scores | Correction efficacy depends on correlation between quality and batch effect |
| Include Batch as Covariate | GLM (in edgeR/DESeq2) | Yes | Simple; integrated into standard DE workflows | Requires balanced design; less effective for strong batch effects |
Experimental Design Considerations
The most effective strategy against batch effects is proactive experimental design. Whenever possible, researchers should ensure that biological conditions of interest are balanced across batches [78]. For instance, in an HCC study comparing tumor to non-tumor tissues, samples from both conditions should be distributed across all sequencing batches. This design enables statistical methods to disentangle biological signals from technical artifacts more effectively.
A critical requirement for successful batch correction is that each batch must contain samples from all biological conditions being studied. The Griffith Lab protocol explicitly warns that "if we processed all the HBR samples with Riboreduction and all the UHR samples with PolyA enrichment, we would be unable to model the batch effect vs the condition effect" [78]. This confounding of batch and condition makes statistical correction impossible.
Principal Component Analysis (PCA) is a valuable diagnostic tool for visualizing batch effects. By plotting samples in the reduced dimension space of the first few principal components and coloring them by both batch and biological condition, researchers can assess whether the primary source of variation is technical (batch) or biological (condition) [78]. A clear separation of batches in the PCA plot indicates a strong batch effect that requires correction.
Integrated Protocol for Batch Effect Correction in HCC ncRNA Studies
Preprocessing and Quality Control
The initial phase focuses on generating high-quality, processed count data suitable for batch correction.
- Raw Read Processing: Begin with raw FASTQ files. Perform adapter trimming and quality filtering using tools such as fastp or Trim Galore [81]. Fastp is noted for its rapid analysis and simplicity, effectively enhancing the quality of processed data [81].
- Alignment and Quantification: Map cleaned reads to an appropriate reference genome (e.g., GRCh38 for human) using a splice-aware aligner like STAR or a pseudo-aligner such as Salmon [80]. For ncRNA-focused studies, ensure the reference includes comprehensive annotations for lncRNAs, miRNAs, and other ncRNAs from databases like miRBase and LNCipedia. Subsequently, generate a raw count matrix at the gene level using tools like featureCounts.
- Quality Assessment: Perform rigorous quality control using FastQC or MultiQC to identify potential technical issues such as adapter contamination, low-quality bases, or unusual base composition [80]. Calculate predicted quality scores (e.g., Plow) if employing quality-aware batch correction methods [79].
- Initial Visualization: Conduct Principal Component Analysis (PCA) on the normalized log-counts-per-million (CPM) values. Color the data points by known batch variables (e.g., sequencing date, library prep kit) and biological conditions (e.g., HCC subtype, tumor vs. non-tumor). This visualization helps assess the severity of batch effects before correction.
Diagram 1: RNA-seq Batch Effect Correction Workflow. This diagram outlines the key steps in a comprehensive RNA-seq analysis pipeline, highlighting stages for quality control, normalization, and batch effect correction.
Batch Correction Using ComBat-ref
For studies involving multiple batches with varying dispersion, ComBat-ref offers superior performance.
- Input Data Preparation: Format the raw count matrix from the quantification step, ensuring that sample batches and conditions are accurately documented.
- Dispersion Estimation: Using the raw counts, employ tools like edgeR to estimate a batch-specific dispersion parameter for each gene within each batch [77].
- Reference Batch Selection: Identify the batch with the smallest dispersion across genes. This batch will serve as the reference to which all other batches will be adjusted [77].
- Model Fitting and Adjustment: Fit a negative binomial GLM to the count data. The model should include terms for the biological condition and batch effects:
log(μ_ijg) = α_g + γ_ig + β_cjg + log(N_j)whereμ_ijgis the expected count for genegin samplejfrom batchi,α_gis the global expression background,γ_igis the effect of batchi,β_cjgis the effect of biological conditionc, andN_jis the library size [77]. The count data from non-reference batches are then adjusted toward the reference batch using the formula:log(μ̃_ijg) = log(μ_ijg) + γ_1g - γ_igwhereγ_1gis the batch effect of the reference batch [77]. - Count Adjustment: The adjusted counts
ñ_ijgare calculated by matching the cumulative distribution function (CDF) of the original negative binomial distribution at the observed countn_ijgwith the CDF of the adjusted distribution atñ_ijg[77]. This ensures the adjusted data retains its count property.
Validation and Downstream Analysis
After applying batch correction, it is essential to validate its effectiveness and proceed with biological interpretation.
- Post-Correction Visualization: Repeat the PCA using the corrected data. A successful correction is indicated by the clustering of samples primarily by biological condition rather than by batch. The Griffith Lab demonstration shows that after proper correction, samples that previously separated by library preparation method (Ribo vs. PolyA) should instead cluster by biological origin (UHR vs. HBR) [78].
- Differential Expression Analysis: Utilize the batch-corrected count data for downstream DE analysis with standard tools like DESeq2 or edgeR. When using ComBat-seq or ComBat-ref, which output adjusted integer counts, these can be directly used in DE tools. If using standard ComBat on normalized data, include the batch as a covariate in the design matrix.
- Biological Validation: Assess the biological plausibility of the results. Compare the list of DE ncRNAs to known pathways implicated in HCC, such as those regulating the tumor immune microenvironment, angiogenesis, epithelial-mesenchymal transition, and metabolism [36]. For instance, confirm whether known oncomiRs like miR-221 or tumor suppressor miRNAs like miR-122 and miR-195 appear appropriately regulated in your HCC samples [2].
Table 3: Key Research Reagent Solutions for RNA-seq Studies in HCC
| Category | Specific Tool/Reagent | Function in Workflow | Application Notes for HCC ncRNA Research |
|---|---|---|---|
| RNA Isolation | miRNeasy Kit (QIAGEN) | Simultaneous purification of total RNA, including small RNAs | Crucial for capturing miRNA and other small ncRNAs from HCC tissue; maintains RNA integrity |
| Library Prep | Ribosomal RNA depletion kits; Poly(A) enrichment | Enrichment for non-ribosomal transcripts or polyadenylated RNA | rRNA depletion captures more lncRNA species; consider library prep method as a potential batch variable |
| cDNA Synthesis | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) | Reverse transcription of RNA into cDNA | Essential for qRT-PCR validation of ncRNA candidates identified by RNA-seq |
| Alignment Reference | GENCODE comprehensive transcriptome; miRBase | Reference for read alignment and quantification | Ensure references include latest lncRNA (e.g., LINC00152, UCA1) and miRNA annotations relevant to HCC |
| Quality Control | FastQC; MultiQC; seqQscorer | Assessment of raw read quality and prediction of sample quality | Low-quality scores can correlate with batch; useful for quality-aware correction methods |
| Batch Correction | ComBat-ref; ComBat-seq (sva R package) | Statistical removal of technical batch variation | ComBat-ref is preferred for multi-batch studies with varying dispersion; preserves count structure |
| Differential Expression | DESeq2; edgeR (R/Bioconductor) | Identification of significantly differentially expressed ncRNAs | Use batch-corrected counts as input; include any residual technical factors in design matrix |
| qRT-PCR Validation | PowerTrack SYBR Green Master Mix; specific primers for ncRNAs | Technical validation of RNA-seq findings for key ncRNAs | Validate findings for key HCC-associated ncRNAs like GAS5, LINC00853 [8] |
Effective management of technical noise through rigorous normalization and advanced batch effect correction is a prerequisite for robust and reproducible RNA-seq analysis, particularly in the complex field of HCC ncRNA research. By integrating the strategies outlined here—from careful experimental design and quality control to the application of specialized tools like ComBat-ref—researchers can significantly enhance the reliability of their data. This approach enables the accurate identification of dysregulated ncRNAs, such as the promising diagnostic panel of LINC00152, LINC00853, UCA1, and GAS5 [8], ultimately advancing our understanding of HCC biology and contributing to the development of much-needed diagnostic and therapeutic strategies.
Navigating Tumor Heterogeneity and Stromal Contamination in Tissue Samples
Hepatocellular carcinoma (HCC) represents a significant global health challenge, characterized by high recurrence rates and poor prognosis. The molecular pathways involved in HCC development are diverse, and no universal molecular feature has been found across all hepatic tumours. A critical obstacle in advancing RNA sequencing analysis of non-coding RNAs in HCC tissues is the inherent tumor heterogeneity and stromal contamination within tissue samples. This complexity is compounded by the tumor microenvironment (TME), a sophisticated ecosystem comprising various cell types including malignant cells, immune cells, and stromal components, alongside noncellular matrix elements. The temporal and spatial interactions among these cells create a complex landscape that profoundly influences clinical outcomes and therapeutic efficacy. Single-cell RNA sequencing (scRNA-seq) has emerged as a transformative technology for investigating specific tumor subtypes, dissecting the complex components of the TME, and elucidating intercellular interactions, thereby providing unprecedented insights into HCC heterogeneity and recurrence mechanisms.
Quantitative Landscape of lncRNAs in HCC
Long non-coding RNAs (lncRNAs), defined as endogenous cellular RNAs longer than 200 nucleotides, have emerged as crucial regulators in physiological and pathological processes, showing differential expression patterns across diverse cancers and affecting their growth and survival potential. In HCC, numerous lncRNAs promote cancer hallmarks including proliferation, invasion, angiogenesis, and migration while inhibiting cellular apoptosis. These functions are mediated through mechanisms such as binding to DNA, RNA, or proteins, inducing epigenetic modifications, or acting as miRNA sponges.
Table 1: Diagnostic Performance of Individual lncRNAs and Machine Learning Models in HCC Detection
| Biomarker | Sensitivity (%) | Specificity (%) | Notes | Citation |
|---|---|---|---|---|
| Individual lncRNAs | 60-83 | 53-67 | Range across LINC00152, LINC00853, UCA1, GAS5 | [8] |
| Machine Learning Model | 100 | 97 | Integrated lncRNAs with conventional laboratory parameters | [8] |
| LINC00152 to GAS5 ratio | N/A | N/A | Significantly correlated with increased mortality risk | [8] |
| lnc-TSPAN12 | N/A | N/A | Independent prognostic predictor for overall and recurrence-free survival | [24] |
RNA-Seq analysis across liver tissues (controls, cirrhotic, and HCC) has revealed compelling data on lncRNA expression. One study detected 5,525 lncRNAs across different tissue types and identified 57 differentially expressed lncRNAs in HCC compared with adjacent non-tumour tissues using stringent criteria (FDR<0.05, Fold Change>2). The number of expressed genes for lncRNAs showed higher variability than protein-coding genes or pseudogenes in each tissue type, with the highest level of variability observed in cirrhotic livers [82].
Table 2: RNA-Sequencing Mapping Statistics for lncRNA Identification in HCC
| Parameter | Value | Details |
|---|---|---|
| Total mapped reads | 857.7 million | Across 23 samples |
| Average reads per sample | 37.3 million | Range: 11.2-72.1 million |
| Percentage mapped to lncRNAs | 6.2% | Of total mapped reads |
| Total expressed genes | 15,172-18,586 | Varies by tissue type (control, cirrhotic, HCC) |
The utilization of lncRNAs as biomarkers is particularly promising because HCC-associated lncRNAs are detectable in body fluids, making them accessible and analyzable, which highlights their potential as valuable biomarkers for liquid biopsy in HCC. Emerging studies indicate that the expression levels of specific lncRNAs in the bloodstream offer promise as non-invasive biomarkers for the early detection and management of HCC [8].
Experimental Protocols for HCC lncRNA Analysis
Protocol 1: Tissue Processing and Single-Cell RNA Sequencing
Principle: This protocol details the procedure for processing HCC tissue samples to perform single-cell RNA sequencing, enabling the dissection of tumor heterogeneity and stromal contamination at cellular resolution.
Materials:
- Fresh HCC tissue samples (primary and relapsed)
- DMEM medium supplemented with 10% FBS
- Collagenase IV (1-2 mg/mL)
- DNase I (0.1-0.5 mg/mL)
- Phosphate Buffered Saline (PBS)
- Red blood cell lysis buffer
- Cell strainers (40μm and 70μm)
- Single-cell RNA sequencing platform (10X Genomics)
- seurat package (v4.4.0) for data analysis
Procedure:
- Tissue Collection: Obtain fresh HCC tissues from surgical resection, ensuring minimal ischemia time (≤30 minutes).
- Tissue Dissociation:
- Mince tissue into 2-4 mm³ fragments using sterile scalpels.
- Transfer to dissociation buffer (Collagenase IV 1-2 mg/mL + DNase I 0.1-0.5 mg/mL) and incubate at 37°C for 30-45 minutes with continuous agitation.
- Cell Suspension Preparation:
- Filter through 70μm cell strainer, followed by 40μm cell strainer.
- Centrifuge at 400 × g for 5 minutes at 4°C.
- Resuspend pellet in red blood cell lysis buffer for 2-3 minutes at room temperature.
- Wash with PBS and resuspend in PBS with 0.04% BSA.
- Cell Quality Control:
- Determine cell viability using Trypan Blue exclusion (>80% viability required).
- Count cells using automated cell counter or hemocytometer.
- Single-Cell RNA Sequencing:
- Load cells onto 10X Genomics Chromium Controller to target 5,000-10,000 cells per sample.
- Prepare libraries according to manufacturer's protocol.
- Sequence on Illumina platform to target 50,000 reads per cell.
- Data Preprocessing:
- Process raw sequencing data using Cell Ranger pipeline.
- Create Seurat object using 'CreateSeuratObject' function.
- Filter cells with 200-10,000 features and genes expressed in at least 10 cells.
- Exclude cells with mitochondrial content exceeding 20% [83].
Protocol 2: lncRNA Quantification via Quantitative Real-Time PCR
Principle: This protocol describes the precise quantification of specific lncRNAs from plasma and tissue samples using quantitative real-time PCR, enabling validation of sequencing results.
Materials:
- Plasma samples or tissue lysates
- miRNeasy Mini Kit (QIAGEN, cat no. 217004)
- RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, cat no. K1622)
- PowerTrack SYBR Green Master Mix kit (Applied Biosystems, cat no. A46012)
- ViiA 7 real-time PCR system (Applied Biosystems)
- Primers for target lncRNAs (LINC00152, LINC00853, UCA1, GAS5)
- GAPDH primer for normalization
Procedure:
- RNA Isolation:
- Extract total RNA using miRNeasy Mini Kit according to manufacturer's protocol.
- Quantify RNA concentration using spectrophotometry (NanoDrop).
- Assess RNA quality (A260/A280 ratio ~2.0).
- cDNA Synthesis:
- Perform reverse transcription using RevertAid First Strand cDNA Synthesis Kit.
- Use 1μg total RNA in 20μL reaction volume.
- Incubate in T100 thermal cycler: 25°C for 5 minutes, 42°C for 60 minutes, 70°C for 5 minutes.
- Quantitative Real-Time PCR:
- Prepare reactions using PowerTrack SYBR Green Master Mix.
- Use 1μL cDNA template in 20μL total reaction volume.
- Run in triplicate on ViiA 7 real-time PCR system with following conditions:
- Hold stage: 95°C for 2 minutes
- PCR stage: 40 cycles of 95°C for 15 seconds and 60°C for 1 minute
- Melt curve stage: 95°C for 15 seconds, 60°C for 1 minute, 95°C for 15 seconds
- Data Analysis:
- Calculate relative expression using ΔΔCT method.
- Normalize to housekeeping gene GAPDH.
- Express results as fold change relative to control group [8].
Protocol 3: Computational Analysis for Heterogeneity Assessment
Principle: This protocol outlines the computational approaches for analyzing scRNA-seq data to decipher tumor heterogeneity and identify lncRNA signatures associated with HCC recurrence.
Materials:
- High-performance computing environment (R version 4.0 or higher)
- R packages: Seurat (v4.4.0), Monocle3, SCENIC, clusterProfiler
- Bulk RNA-seq data from TCGA (https://cancergenome.nih.gov/)
- Validation datasets from GEO (accession GSE14520)
Procedure:
- Data Integration:
- Merge multiple scRNA-seq datasets using harmony or Seurat's integration workflow.
- Normalize data using SCTransform method.
- Scale and center features for dimensionality reduction.
- Cell Type Identification:
- Perform principal component analysis (PCA) on variable features.
- Apply graph-based clustering (resolution 0.4-0.8).
- Generate UMAP/t-SNE plots for visualization.
- Annotate cell types using known marker genes.
- Differential Expression Analysis:
- Identify differentially expressed lncRNAs using Wilcoxon rank sum test.
- Apply Bonferroni correction for multiple testing (adjusted p-value < 0.05).
- Calculate average log2 fold change (threshold > 0.25).
- Trajectory Inference:
- Construct single-cell trajectories using Monocle3.
- Order cells along pseudotime to infer differentiation pathways.
- Identify lncRNAs associated with state transitions.
- Machine Learning Model Development:
- Construct relapsed tumor cell-related risk score (RTRS) using multiple machine learning methods.
- Validate prognostic accuracy using TCGA and GEO datasets.
- Compare predictive performance with conventional clinical variables [83].
Visualizing Experimental Workflows and Signaling Pathways
Experimental Workflow for HCC Heterogeneity Analysis
MIF Signaling in HCC Recurrence
Research Reagent Solutions
Table 3: Essential Research Reagents for HCC lncRNA Studies
| Reagent/Kit | Manufacturer | Function | Application Note |
|---|---|---|---|
| miRNeasy Mini Kit | QIAGEN (cat no. 217004) | Total RNA isolation from cells and tissues | Optimal for simultaneous recovery of long and short RNAs |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific (cat no. K1622) | Reverse transcription of RNA to cDNA | Includes RNase H+ M-MuLV Reverse Transcriptase for full-length cDNA |
| PowerTrack SYGR Green Master Mix | Applied Biosystems (cat no. A46012) | qRT-PCR amplification and detection | Optimized for difficult templates with high GC content |
| Collagenase IV | Various suppliers | Tissue dissociation for single-cell preparations | Concentration 1-2 mg/mL with 30-45 minute incubation |
| 10X Genomics Chromium Controller | 10X Genomics | Single-cell partitioning and barcoding | Targets 5,000-10,000 cells per sample |
| Seurat R Package | CRAN (v4.4.0) | Single-cell data analysis and visualization | Comprehensive toolkit for scRNA-seq analysis |
Discussion and Future Perspectives
The integration of single-cell technologies with computational approaches represents a paradigm shift in navigating tumor heterogeneity and stromal contamination in HCC research. Machine learning models that integrate lncRNA expression profiles with conventional clinical parameters have demonstrated remarkable diagnostic performance, achieving up to 100% sensitivity and 97% specificity in HCC detection [8]. The development of relapsed tumor cell-related risk score (RTRS) models using multiple machine learning methods has shown higher accuracy in predicting overall and recurrence-free survival compared with conventional clinical variables [83].
Future directions should focus on the standardization of analytical protocols, enhancement of multi-omics integration, and development of more sophisticated computational models that can fully leverage the complexity of single-cell data. Furthermore, the clinical translation of these findings requires robust validation in prospective cohorts and the development of accessible diagnostic platforms that can implement these complex analyses in routine clinical practice. As our understanding of HCC heterogeneity deepens, personalized therapeutic strategies targeting specific lncRNA networks and cellular subpopulations will become increasingly feasible, ultimately improving outcomes for patients with this devastating disease.
Optimizing Feature Selection and Dimensionality Reduction for High-Dimensional Data
The analysis of non-coding RNAs (ncRNAs) in hepatocellular carcinoma (HCC) tissues represents a classic high-dimensional data challenge, where the number of features (ncRNA transcripts) vastly exceeds the number of patient samples. This "curse of dimensionality" is particularly pronounced in RNA sequencing studies, where researchers routinely measure the expression of thousands of miRNAs, lncRNAs, and circRNAs from limited clinical specimens. Feature selection and dimensionality reduction have therefore become indispensable preprocessing steps for building robust, interpretable, and clinically applicable models in HCC research [84] [85].
The biological complexity of the non-coding transcriptome necessitates sophisticated computational approaches. Unlike traditional statistical methods that consider limited interactions, machine learning (ML) algorithms can identify complex, nonlinear relationships between ncRNAs and clinical outcomes, capturing the multifaceted interplay inherent in ncRNA regulatory networks [84]. This application note provides a structured framework for optimizing these critical computational steps specifically within the context of HCC ncRNA research.
Foundational Concepts and Methodologies
Distinguishing Feature Selection from Dimensionality Reduction
In the context of ncRNA data analysis, it is crucial to understand the distinction between two complementary approaches:
- Feature Selection: Identifies and retains a subset of the most relevant ncRNA biomarkers from the original feature space. This preserves the biological interpretability of the selected miRNAs, lncRNAs, or circRNAs, which is essential for understanding their mechanistic roles in hepatocarcinogenesis [85].
- Dimensionality Reduction: Transforms the high-dimensional ncRNA expression data into a lower-dimensional space using feature extraction techniques. This creates new, composite features (e.g., principal components) that may enhance model performance but can obscure direct biological interpretation [86].
Taxonomy of Feature Selection Methods for ncRNA Data
Table 1: Categories of Feature Selection Methods Applicable to ncRNA Studies
| Method Category | Core Principle | Advantages | Disadvantages | Example Algorithms |
|---|---|---|---|---|
| Filter Methods | Selects features based on statistical measures independent of ML model. | Computationally fast, scalable, resistant to overfitting. | Ignores feature dependencies, may select redundant features. | Signal-to-Noise Ratio (SNR), Mood's median test [87], χ² test [85]. |
| Wrapper Methods | Uses the performance of a predictive model to evaluate feature subsets. | Captures feature interactions, often high-performing. | Computationally intensive, risk of overfitting. | Recursive Feature Elimination, Binary Black Particle Swarm Optimization (BBPSO) [88]. |
| Embedded Methods | Performs feature selection as part of the model construction process. | Balances efficiency and performance, models feature interactions. | Model-specific selection. | LASSO regression [89], Random Forest feature importance [90] [85]. |
Advanced Hybrid and Ensemble Approaches
Recent advances leverage hybrid frameworks that combine the strengths of multiple paradigms. For instance, one study introduced a method combining the Signal-to-Noise Ratio (SNR) score with the robust Mood median test to identify genes with significant changes across groups while reducing the impact of outliers common in skewed biological data [87]. Similarly, hybrid metaheuristic algorithms like Two-phase Mutation Grey Wolf Optimization (TMGWO) and Improved Salp Swarm Algorithm (ISSA) have demonstrated superior performance in selecting optimal feature subsets from high-dimensional datasets, achieving high classification accuracy for disease diagnosis [88].
Performance Benchmarking in HCC Context
Empirical evidence from recent HCC studies demonstrates the tangible benefits of optimized feature selection. The performance gains are consistent across various ML algorithms and ncRNA types.
Table 2: Performance Comparison of ML Models with Feature Selection in HCC Diagnostics
| Study Focus | Selected Features | ML Algorithm(s) | Key Performance Metrics | Reference |
|---|---|---|---|---|
| HCC Diagnosis | RAB11A, STAT1, ATG12, miR-1262, miR-1298, miR-106b-3p, lncRNA-RP11-513I15.6, lncRNA-WRAP53, plus clinical data | LGBM, Random Forest, DNN, SVC, KNN | LGBM achieved highest accuracy: 98.75% [90] | [90] |
| HCC Screening | LINC00152, LINC00853, UCA1, GAS5 lncRNAs combined with conventional lab parameters | Machine Learning Model (Python Scikit-learn) | Sensitivity: 100%, Specificity: 97% [8] | [8] |
| General High-Dim. Data Classification | Various feature subsets selected via hybrid algorithms | TMGWO with SVM | Achieved 96% accuracy on Breast Cancer dataset using only 4 features [88] | [88] |
Experimental Protocols and Workflows
Integrated Protocol for ncRNA Feature Selection and Model Building
This protocol outlines a comprehensive workflow from raw ncRNA data to a validated predictive model, incorporating best practices for feature selection.
Phase 1: Data Preprocessing and Quality Control
- RNA Extraction & QC: Purify total RNA from serum/plasma or HCC tissues using a miRNEasy extraction kit (Qiagen). Validate RNA quality and purity using a fluorimeter (e.g., Qubit 3.0) [90].
- Reverse Transcription: Convert RNA to cDNA using a dedicated kit (e.g., miScript II RT Kit) [90].
- Expression Quantification: Perform quantitative RT-PCR (qRT-PCR) for specific ncRNAs of interest using SYBR Green-based chemistry. For discovery workflows, use RNA-sequencing or microarray platforms [90] [70].
- Data Normalization: Calculate relative expression (RQ) using the 2−ΔΔCt method with appropriate endogenous controls (e.g., SNORD72 for miRNAs, GAPDH for mRNAs) [90].
- Quality Filtering: Remove low-quality samples and low-expression ncRNAs. Apply variance-based filtering to remove non-informative features.
Phase 2: Feature Selection Execution
- Initial Filtering: Apply a univariate filter method (e.g., SNR with Mood median test [87]) to reduce the feature space by 50-60%.
- Redundancy Analysis: Account for linkage disequilibrium or highly correlated ncRNAs to select a representative feature from correlated clusters [85].
- Advanced Selection: Implement a wrapper or embedded method (e.g., Random Forest feature importance or LASSO) on the filtered subset to identify the final panel of ncRNA biomarkers.
Phase 3: Model Building and Validation
- Algorithm Selection: Train multiple ML classifiers (e.g., LGBM, RF, SVM) using the selected features.
- Hyperparameter Tuning: Optimize algorithm-specific parameters via grid or random search.
- Rigorous Validation: Perform K-fold cross-validation (e.g., K=5 or 10) to assess performance and avoid overfitting [85]. Test the final model on an independent validation cohort.
Diagram 1: Integrated ncRNA Analysis Workflow (Width: 760px)
Protocol for Dimensionality Reduction of scRNA-Seq Data
Single-cell RNA sequencing (scRNA-Seq) of HCC tissues presents additional challenges of extreme dimensionality and data sparsity (dropout events). The following protocol is adapted for scRNA-Seq data:
- Post-Sequencing Processing: Process FASTQ files through a standardized pipeline (e.g., Cell Ranger for 10x Genomics data) to obtain a gene expression (UMI) matrix [86].
- Normalization: Normalize the raw count data to account for sequencing depth variability.
- Highly Variable Feature Identification: Identify genes that exhibit high cell-to-cell variation, which are likely to contain biologically relevant information.
- Dimensionality Reduction via PCA: Perform Principal Component Analysis (PCA) on the scaled data of highly variable genes. Select the top principal components (PCs) that capture the majority of the biological variance, typically using the 'elbow' method [86].
- Non-linear Visualization: Use the top PCs as input for non-linear visualization techniques like t-SNE or UMAP to project cells into 2D/3D space for cluster identification and exploratory analysis [86].
- Downstream Analysis: Proceed with cell clustering, differential expression analysis, and cell-type annotation based on the reduced-dimensional space.
Table 3: Key Reagent Solutions for ncRNA Feature Selection Workflows
| Reagent / Tool | Specific Example | Function in Workflow | Reference |
|---|---|---|---|
| RNA Extraction Kit | miRNEasy Mini Kit (Qiagen, cat no. 217004) | Purifies total RNA (including small RNAs) from serum, plasma, or tissues. | [90] [8] |
| cDNA Synthesis Kit | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) | Reverse transcribes RNA into cDNA for subsequent qPCR analysis. | [8] |
| qRT-PCR Master Mix | Quantitect SYBR Green, miScript SYBR Green PCR Kit | Enables quantification of specific ncRNA transcript levels. | [90] |
| NGS Platform | 10x Genomics Chromium (for scRNA-Seq) | Provides high-throughput transcriptomic data at single-cell resolution. | [86] |
| Feature Selection Software | Python Scikit-learn, R Caret package | Provides implementations of filter, wrapper, and embedded feature selection methods. | [88] [8] |
| ML Algorithms | LightGBM (LGBM), Random Forest, SVM | High-performance classifiers for building diagnostic/prognostic models. | [90] [88] |
Case Study: Implementing a Diagnostic Model for HCC
A practical example from the literature illustrates this workflow. A study aimed to develop a diagnostic model for HCC using five different classifiers (KNN, RF, SVM, LGBM, DNN). The model incorporated 22 features, including key ncRNAs (RQmiR-1298, RQmiR-1262, RQmRNARAB11A, RQSTAT1, RQLnc-WRAP53, etc.) and clinical parameters (age, sex, AFP, ALT, AST) [90].
- Feature Selection Implicitly Applied: While not explicitly detailing their feature selection method, the study's model was built on a curated set of ncRNAs previously identified as significant, representing a knowledge-driven pre-selection step [90].
- Performance Outcome: The LGBM algorithm achieved the highest accuracy of 98.75% on the test set, demonstrating the power of combining a selected panel of ncRNA biomarkers with a robust ML algorithm [90].
Diagram 2: HCC Diagnostic Model Pipeline (Width: 760px)
Optimizing feature selection and dimensionality reduction is not merely a computational exercise but a critical component in translating ncRNA discoveries into clinically actionable insights for HCC. The structured protocols and benchmarks provided here offer a roadmap for researchers to enhance the reliability and performance of their models. Future directions will likely involve the deeper integration of multi-omics data, the application of explainable AI (XAI) to interpret complex models, and the development of feature selection methods specifically designed to handle the unique characteristics of single-cell and long-read RNA sequencing technologies. By rigorously applying these principles, the path from high-dimensional ncRNA data to robust diagnostic, prognostic, and therapeutic biomarkers in HCC can be significantly accelerated.
Best Practices for Functional Enrichment Analysis of ncRNA Targets and Pathways
Within the context of hepatocellular carcinoma (HCC) research, functional enrichment analysis has emerged as a critical bioinformatics process for translating lists of differentially expressed non-coding RNAs (ncRNAs) into mechanistically meaningful biological insights. HCC is a complex and lethal malignancy characterized by heterogeneous molecular alterations, where ncRNAs—including long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs)—have been shown to play essential regulatory roles in tumor initiation, progression, and metastasis [3] [24]. Traditional functional annotation methods that focus solely on direct connections between ncRNAs and protein-coding genes (PCGs) often overlook the global crosstalk within biomolecular networks, limiting their applicability and accuracy [91]. This Application Note establishes a comprehensive framework for conducting robust functional enrichment analysis specifically tailored to ncRNA studies in HCC, integrating advanced computational approaches with practical experimental validation strategies to elucidate the pathways and processes driven by ncRNA dysregulation in liver carcinogenesis.
A Comprehensive Framework for ncRNA Functional Annotation
The ncFN Framework: An Integrated Approach
The ncFN (non-coding RNA Function annotation) framework represents a significant advancement in ncRNA functional analysis by leveraging a Global Interaction Network (GIN) that encompasses heterogeneous interactions between multiple types of ncRNAs and PCGs [91]. This approach addresses a critical limitation of conventional methods that typically focus on a single ncRNA type by integrating:
- PCG-PCG interactions from pathway databases (Reactome, KEGG, NetPath), protein-protein interactions (PPIs), and transcription factor (TF)-target gene pairs
- ncRNA-PCG interactions including lncRNA-PCG (from starBase, LncRNA2Target), miRNA-PCG (from mirTarBase), and TF-miRNA (from TransmiR) relationships
- ncRNA-ncRNA interactions such as miRNA-lncRNA (from LncBase) and miRNA-circRNA (from starBase) associations [91]
The assembled GIN in ncFN consists of 565,482 edges connecting 17,060 PCGs and 12,616 ncRNAs (including 1,095 miRNAs, 3,563 lncRNAs, and 7,958 circRNAs), providing an extensive foundation for comprehensive functional analysis [91].
Association Strength Quantification Using Random Walk with Restart
For each ncRNA of interest, ncFN quantifies Association Strengths (ASs) between the ncRNA and PCGs through Random Walk with Restart (RWR) analysis on the GIN [91]. The RWR algorithm simulates a walker that traverses the network, starting at the seed ncRNA node, and iteratively computes relevance scores for all other nodes based on network connectivity. The mathematical formulation is:
Pₜ₊₠= (1-r)WPₜ + rP₀
Where Pâ‚€ represents the initial probability vector (with value 1 for the seed node), Pâ‚œ denotes the probability distribution at iteration step t, W is the column-normalized adjacency matrix, and r is the restart coefficient governing the balance between local exploration and global diffusion [91]. The algorithm iterates until convergence (|Pₜ₊₠- Pâ‚œ| < 10â»Â¹â°), with the resulting stationary probability distribution defining the ASs between all nodes and the seed ncRNA.
Functional Annotation via Gene Set Enrichment Analysis
Following AS calculation, pre-ranked Gene Set Enrichment Analysis (GSEA) is performed using PCGs ranked by their ASs as an ordered gene list against a collection of functional gene sets (typically 299 KEGG pathways) [91] [92]. This approach identifies pathways enriched among PCGs with the strongest network associations to the query ncRNA, providing a systems-level understanding of its potential biological functions beyond direct targets.
Table 1: Key Resources for ncRNA Functional Enrichment Analysis
| Resource Type | Specific Tools/Databases | Primary Function | Application Context |
|---|---|---|---|
| Pathway Databases | KEGG, Reactome, WikiPathways, PANTHER, NetPath | Provide curated gene sets representing biological pathways | Functional gene sets for enrichment testing [92] |
| Interaction Databases | starBase, LncRNA2Target, mirTarBase, LncBase, TransmiR | Experimentally validated ncRNA-RNA/protein interactions | Building molecular networks for association analysis [91] |
| Enrichment Tools | g:Profiler, GSEA, clusterProfiler, EnrichmentMap | Perform statistical enrichment analysis and visualization | Identifying significantly overrepresented pathways [92] [93] |
| Network Analysis | Cytoscape, STRING, ncFN | Network construction, analysis, and visualization | Exploring complex molecular relationships [91] [94] |
Experimental Protocols and Workflows
Protocol 1: ncRNA Functional Annotation Using the ncFN Framework
Step 1: Data Preparation and Integration
- Collect ncRNA expression data from HCC tissues versus normal adjacent tissues using RNA-seq or single-cell RNA-seq
- Identify significantly dysregulated ncRNAs (FDR < 0.05, |logâ‚‚FC| > 1) using tools like edgeR or DESeq2
- Annotate ncRNAs using authoritative databases (miRBase for miRNAs, GENCODE for lncRNAs, circBase for circRNAs) [91]
Step 2: Global Interaction Network Construction
- Download molecular interactions from curated databases (see Table 1)
- Standardize molecular identifiers (Entrez Gene IDs for PCGs, Ensembl IDs for lncRNAs, miRBase accessions for miRNAs, circBase IDs for circRNAs)
- Construct the integrated network and extract the largest connected component using igraph R package (v1.4.0) [91]
- Treat all edges as unweighted and undirected to comprehensively capture regulatory relationships
Step 3: Association Strength Calculation
- Implement Random Walk with Restart algorithm using the ncRNA of interest as seed node
- Set restart parameter (r) between 0.1-0.9 (optimal value can be determined by testing how effectively known disease ncRNAs prioritize their established pathways)
- Run iterations until convergence (difference between Pₜ₊₠and Pâ‚œ < 10â»Â¹â°)
- Extract the final probability distribution as Association Strengths for all PCGs [91]
Step 4: Functional Enrichment Analysis
- Rank all PCGs by their Association Strengths from highest to lowest
- Perform pre-ranked GSEA against KEGG pathway gene sets using standard parameters
- Apply significance thresholds (NOM p-value < 0.05, FDR q-value < 0.25) to identify enriched pathways [91]
- For HCC studies, pay particular attention to pathways commonly dysregulated in cancer (e.g., cell cycle, apoptosis, metabolic pathways)
Step 5: Results Interpretation and Validation
- Visualize enrichment results using bar plots, dot plots, or enrichment maps [93]
- Cross-reference with known HCC pathways and previously established ncRNA functions
- Select top candidate pathways for experimental validation using in vitro and in vivo models
Diagram 1: ncRNA Functional Annotation Workflow using the ncFN framework. RWR = Random Walk with Restart; GSEA = Gene Set Enrichment Analysis.
Protocol 2: ceRNA Network Analysis in HCC
The competing endogenous RNA (ceRNA) hypothesis proposes that different RNA transcripts can communicate through shared miRNA response elements, forming a complex regulatory network particularly relevant in cancer biology including HCC [94].
Step 1: Identification of Differentially Expressed RNAs
- Isolate RNA from HCC tissues and matched normal tissues
- Perform RNA-seq for comprehensive profiling of lncRNAs, miRNAs, and mRNAs
- Calculate expression levels (FPKM for lncRNAs/mRNAs, TPM for miRNAs)
- Identify differentially expressed RNAs using DEGseq (miRNAs) and edgeR (lncRNAs/mRNAs) with threshold FDR < 0.01 and |logâ‚‚FC| > 1 [94]
Step 2: miRNA Target Prediction
- Predict miRNA-mRNA interactions using multiple algorithms: miRanda (min free energy ≤ -10 kcal/mol), PITA, and RNAhybrid
- Retain only interactions identified by all three tools to increase confidence
- Predict miRNA-lncRNA pairs using miRanda [94]
Step 3: ceRNA Network Construction
- Select lncRNA-miRNA and miRNA-mRNA pairs showing negative correlation of expression
- Identify lncRNA-mRNA pairs with positive correlation of expression and shared miRNA binding sites
- Integrate DEmiRNA-DEmRNA and DEmiRNA-DElncRNA pairs into the ceRNA network
- Visualize the network using Cytoscape (v3.5.0 or higher) [94]
Step 4: Functional Enrichment of ceRNA Components
- Extract protein-coding genes from the ceRNA network
- Perform KEGG pathway enrichment analysis using KOBAS software (v2.0)
- Conduct Gene Ontology enrichment analysis using GOseq R package [94]
- Focus on HCC-relevant pathways identified through enrichment (e.g., NF-kappa B signaling, cell cycle, apoptosis)
Step 5: Experimental Validation of Key ceRNA Interactions
- Validate top ceRNA interactions using luciferase reporter assays
- Perform functional studies through knockdown/overexpression of network components
- Assess impact on HCC phenotypes (proliferation, migration, invasion)
Table 2: Example ceRNA Network Analysis Results from Down Syndrome Study (Illustrative Methodology)
| Network Component | Upregulated RNAs | Downregulated RNAs | Key Pathways Identified |
|---|---|---|---|
| DEmiRNAs | 88 miRNAs | 128 miRNAs | - |
| DElncRNAs | 154 transcripts | 497 transcripts | - |
| DEmRNAs | 3,915 transcripts | 7,818 transcripts | NF-kappa B signaling, T-cell receptor signaling, Apoptosis |
| Hub Genes in PPI | RPS27A, UBA52, UBC, RPL11, RPS27 | NFKB1, RBX1, RELA | Ribosome, Oxidative phosphorylation, Alzheimer's disease |
Visualization and Interpretation of Results
Effective visualization is essential for interpreting functional enrichment results and communicating findings. Multiple complementary approaches should be employed:
Enrichment Map Visualization
- Construct enrichment maps using Cytoscape with the EnrichmentMap plugin to organize enriched terms into a network
- Connect overlapping gene sets to identify functional modules
- Use node size to represent the number of genes in each term and edge thickness to represent overlap between terms [93]
Gene-Concept Network Diagrams
- Generate gene-concept networks (cnetplots) to depict linkages between genes and biological concepts using the enrichplot package in R
- Display complex associations where genes may belong to multiple annotation categories
- For GSEA results, display core enriched genes to highlight key contributors to pathway enrichment [93]
Heatmap-like Functional Classification
- Create heatplots to simplify visualization when dealing with large numbers of significant terms
- Display relationships between genes and pathways as a heatmap to identify expression patterns more easily than complex networks [93]
Tree Plot Hierarchical Clustering
- Perform hierarchical clustering of enriched terms using treeplot() function based on pairwise similarities (Jaccard's similarity index or semantic similarity)
- Cut the tree into subtrees (default: 5 clusters) and label using high-frequency words to reduce complexity and improve interpretation [93]
Diagram 2: Visualization Strategies for Functional Enrichment Results.
Table 3: Research Reagent Solutions for ncRNA Functional Studies
| Reagent/Resource | Function | Example Applications | Key Features |
|---|---|---|---|
| Trizol Reagent | RNA isolation from tissues/cells | Extract total RNA from HCC tissues and matched normals | Maintains RNA integrity, suitable for multiple RNA types |
| edgeR/DEGseq | Differential expression analysis | Identify DE ncRNAs from RNA-seq data | Handles count data with dispersion estimation, precise for small samples |
| clusterProfiler | Functional enrichment analysis | ORA and GSEA for ncRNA target genes | Integrates with bioconductor, supports multiple organisms |
| Cytoscape | Network visualization and analysis | ceRNA network construction and analysis | Plugin ecosystem, handles large networks |
| STRING Database | Protein-protein interaction data | PPI network for ncRNA target genes | Confidence scores, comprehensive coverage |
| mirTarBase | Experimentally validated miRNA targets | miRNA-mRNA interaction evidence | Quality ratings, multi-species support |
Functional enrichment analysis of ncRNA targets and pathways represents a powerful approach for elucidating the mechanistic roles of ncRNAs in HCC pathogenesis. The integration of comprehensive molecular networks, robust statistical methods for association scoring, and sophisticated visualization techniques enables researchers to move beyond simple differential expression to functional understanding. The protocols and best practices outlined here provide a structured framework for applying these approaches to HCC research, with the ultimate goal of identifying novel diagnostic biomarkers, therapeutic targets, and biological insights for this devastating malignancy. As single-cell technologies and spatial transcriptomics continue to advance, these functional analysis methods will become increasingly important for understanding ncRNA functions at cellular resolution in the complex tumor microenvironment of hepatocellular carcinoma.
The integration of high-throughput RNA sequencing (RNA-seq) technologies and sophisticated in-silico prediction tools has revolutionized the discovery of novel biomarkers and therapeutic targets in complex diseases like hepatocellular carcinoma (HCC). However, a significant validation gap often exists between computational predictions and their biological confirmation, particularly in the rapidly evolving field of non-coding RNA (ncRNA) research in HCC tissues. This application note provides a structured framework and detailed protocols to bridge this gap, enabling researchers to effectively translate computational discoveries into experimentally validated findings with clinical translational potential. We focus specifically on the context of ncRNA research in HCC, where molecules like long non-coding RNAs (lncRNAs) play critical regulatory roles in tumor proliferation, metastasis, and apoptosis [28] [95].
Key Research Reagent Solutions for HCC ncRNA Studies
The table below summarizes essential reagents and databases critical for conducting integrated in-silico and experimental studies on ncRNAs in HCC.
Table 1: Essential Research Reagents and Databases for HCC ncRNA Research
| Category | Specific Resource/Reagent | Function/Application | Example Use Case in HCC |
|---|---|---|---|
| Public Data Repositories | GEO/SRA [96] | Access to raw and processed RNA-seq data; hypothesis generation and validation | Download HCC and non-tumor liver tissue datasets (e.g., GSE101685, GSE14520) for differential expression analysis [97]. |
| Public Data Repositories | TCGA (The Cancer Genome Atlas) [96] | Repository for cancer genomics data, including clinical information | Obtain HCC patient RNA-seq data (TCGA-LIHC) to correlate ncRNA expression with survival and other clinical parameters [97]. |
| Public Data Repositories | ARCHS4 / Recount3 [96] | Resource of uniformly processed RNA-seq data from public sources | Rapidly access and analyze a large number of HCC-related gene expression samples for meta-analysis. |
| Bioinformatics Tools | "Limma" R package [97] | Statistical analysis for identifying differentially expressed genes (DEGs) | Identify lncRNAs and other ncRNAs significantly dysregulated in HCC tissues compared to normal controls. |
| Bioinformatics Tools | "WGCNA" R package [97] | Weighted Gene Co-expression Network Analysis to find gene modules | Discover co-expressed ncRNA-mRNA networks associated with specific HCC clinical traits or pathways. |
| Bioinformatics Tools | "clusterProfiler" R package [97] | Functional enrichment analysis (GO, KEGG) | Interpret the biological roles and pathways of predicted HCC-associated ncRNAs. |
| Experimental Reagents | HepG2.2.15 cell line [76] | HBV-infected hepatoblastoma cell line for modeling virus-associated HCC | Study the role of specific lncRNAs (e.g., in immune response pathways like SERPINA1) in an HBV context [76]. |
| Experimental Reagents | scRNA-seq Platform (e.g., 10x Genomics) | Profiling transcriptional heterogeneity at single-cell resolution | Characterize distinct cell subpopulations and ncRNA expression within the HCC tumor microenvironment [76]. |
Integrated Workflow: From Prediction to Validation
The following diagram outlines a comprehensive, multi-stage workflow designed to bridge the validation gap in HCC ncRNA research, from initial bioinformatics discovery to final functional confirmation.
Detailed Experimental Protocols
Protocol: In-Silico Identification of HCC-Associated ncRNAs
Objective: To identify differentially expressed long non-coding RNAs (lncRNAs) from public HCC RNA-seq datasets using a standardized bioinformatics pipeline.
Materials:
- R statistical software environment (v4.0 or higher)
- R packages:
limma,DESeq2,clusterProfiler,WGCNA,edgeR[97] [60] - Public dataset accession numbers (e.g., GSE101685, GSE14520, TCGA-LIHC from GEO and TCGA portals) [97] [96]
Procedure:
- Data Acquisition and Preprocessing:
- Download raw count data or processed expression matrices for HCC and normal liver tissues from chosen datasets in public repositories like GEO or TCGA [96].
- Perform quality control checks. Normalize raw count data using appropriate methods (e.g., TMM for edgeR, variance stabilizing transformation for DESeq2) to account for library size and composition biases [60].
- Correct for batch effects using the
ComBatfunction from thesvapackage in R [97].
Differential Expression Analysis:
- Using the
limmapackage, fit a linear model to the normalized expression data to identify genes and lncRNAs significantly differentially expressed between HCC and normal samples [97]. - Apply a false discovery rate (FDR) correction (e.g., Benjamini-Hochberg). Set significance thresholds (e.g., |logFC| > 1, FDR-adjusted p-value < 0.05) [97].
- Generate a volcano plot for visualization using
ggplot2.
- Using the
Co-expression Network Analysis (WGCNA):
- Construct a weighted gene co-expression network using the
WGCNApackage on the normalized expression data [97]. - Identify modules of highly co-expressed genes. Correlate module eigengenes with clinical traits of interest (e.g., tumor stage, survival) to select biologically relevant modules.
- Extract lncRNAs within significant modules for further analysis.
- Construct a weighted gene co-expression network using the
Functional Enrichment Analysis:
- Perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on protein-coding genes that are co-expressed with the candidate lncRNAs or within the same WGCNA module using
clusterProfiler[97]. - This step helps infer the potential biological roles and pathways in which the candidate lncRNAs may be involved.
- Perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on protein-coding genes that are co-expressed with the candidate lncRNAs or within the same WGCNA module using
Output: A prioritized list of candidate lncRNAs with significant differential expression, association with clinically relevant modules, and inferred functional roles.
Protocol: Single-Cell RNA-seq Validation of Candidate ncRNAs
Objective: To validate the expression and explore the cellular distribution of candidate ncRNAs at single-cell resolution within the HCC tumor microenvironment (TME).
Materials:
- Fresh or frozen HCC tissue specimens and matched non-tumor liver tissue.
- Single-cell RNA-seq platform (e.g., 10x Genomics).
- Computational resources and software (e.g., Seurat, SingleCellExperiment in R) [76].
Procedure:
- Sample Preparation and Sequencing:
- Generate single-cell suspensions from dissociated HCC and control tissues.
- Proceed with library preparation using a platform-specific kit (e.g., 10x Genomics 3' Gene Expression).
- Sequence the libraries on an appropriate Illumina platform to a recommended depth (e.g., 50,000 reads per cell).
Data Preprocessing and Quality Control:
- Process raw sequencing data (demultiplexing, alignment, and gene counting) using the platform's pipeline (e.g.,
cellranger). - Import the data into an R environment and filter out low-quality cells using the following criteria [76]:
- Cells with fewer than 200 or more than 2500 detected genes.
- Cells with >5% mitochondrial read count, indicating stress or apoptosis.
- Normalize the data to account for sequencing depth and log-transform.
- Process raw sequencing data (demultiplexing, alignment, and gene counting) using the platform's pipeline (e.g.,
Feature Selection, Dimensionality Reduction, and Clustering:
- Identify 2000-3000 highly variable genes (HVGs) that drive heterogeneity.
- Perform principal component analysis (PCA) on the HVGs.
- Use the top principal components (determined by an elbow plot) to cluster cells with a graph-based algorithm (e.g., Louvain) and project cells into two dimensions using UMAP [76].
Cell Type Annotation and Candidate ncRNA Validation:
- Annotate cell clusters using known marker genes (e.g., ALB for hepatocytes, PECAM1 for endothelial cells, CD68 for macrophages) or a reference-based annotation tool like
SingleR[76]. - Visualize the expression level of your candidate lncRNAs across the annotated cell types on the UMAP plot and using violin plots.
- Confirm specific expression or significant up/down-regulation of the candidate in relevant cell populations (e.g., malignant hepatocytes, specific immune subsets) of the HCC sample compared to the normal control.
- Annotate cell clusters using known marker genes (e.g., ALB for hepatocytes, PECAM1 for endothelial cells, CD68 for macrophages) or a reference-based annotation tool like
Output: Validated cell-type-specific expression patterns of candidate ncRNAs, providing insight into their potential functional context within the heterogeneous HCC TME.
Protocol: Functional Validation of ncRNAs Using In Vitro Models
Objective: To assess the functional impact of a candidate lncRNA on HCC cell phenotypes through gain- and loss-of-function experiments.
Materials:
- Human HCC cell lines (e.g., HepG2, Huh-7, HepG2.2.15 for HBV-related studies [76]).
- siRNAs or ASOs targeting the candidate lncRNA; cDNA overexpression construct for the lncRNA.
- Transfection reagent.
- qRT-PCR reagents for validation of knockdown/overexpression.
- Assay kits for cell viability (e.g., MTS, CCK-8), migration (e.g., transwell), and apoptosis (e.g., caspase-3/7 activity).
Procedure:
- Modulate Candidate lncRNA Expression:
- Seed HCC cells in appropriate culture plates and transfert with:
- Knockdown Group: siRNA or antisense oligonucleotides (ASOs) targeting the lncRNA.
- Overexpression Group: Plasmid construct overexpressing the full-length lncRNA.
- Control Groups: Non-targeting siRNA/scrambled ASO or empty vector.
- Incubate for 24-72 hours.
- Seed HCC cells in appropriate culture plates and transfert with:
Confirm Knockdown/Overexpression Efficiency:
- Extract total RNA from all treatment groups.
- Synthesize cDNA and perform qRT-PCR using primers specific for the candidate lncRNA.
- Normalize expression to a housekeeping gene (e.g., GAPDH, ACTB). Confirm significant knockdown (>70%) or overexpression relative to controls.
Phenotypic Assays:
- Cell Proliferation/Viability: Seed transfected cells in 96-well plates. Measure viability at 0, 24, 48, and 72 hours using an MTS or CCK-8 assay according to the manufacturer's protocol.
- Cell Migration: 24 hours post-transfection, seed cells into transwell inserts (without serum). Place inserts in plates with medium containing serum as a chemoattractant. After 24-48 hours, fix, stain, and count migrated cells on the lower membrane surface.
- Apoptosis Assay: 48 hours post-transfection, measure caspase-3/7 activity using a luminescent assay kit as per the manufacturer's instructions.
Output: Quantitative data linking the candidate lncRNA's expression level to key oncogenic phenotypes (proliferation, migration, apoptosis resistance) in HCC cells.
Data Presentation and Analysis
The quantitative data generated from the aforementioned protocols should be systematically analyzed and presented. The table below provides a template for summarizing key findings from the differential expression and initial validation stages.
Table 2: Summary Table for Candidate HCC-associated ncRNA Validation Data
| Candidate ncRNA | In-Silico Analysis (Bulk RNA-seq) | scRNA-seq Validation | Functional Validation (Phenotype) | |||
|---|---|---|---|---|---|---|
| Log2FC | Adj. p-value | Primary Expressing Cell Type | Expression in HCC | Proliferation (vs Ctrl) | Migration (vs Ctrl) | |
| LINC01134 | +3.5 | 1.2e-08 | Malignant Hepatocytes | Up | +40% * | +55% * |
| FAM111A-DT | +4.1 | 5.8e-10 Cancer-Associated Fibroblasts | Up | No significant change | +25% | |
| CERS6-AS1 | -2.8 | 3.4e-06 Endothelial Cells | Down | -15% * | -20% | |
| NEAT1 | +2.2 | 7.1e-05 Multiple Immune Cells | Up | +30% | +35% |
Note: Data in this table is illustrative. FC: Fold Change; Ctrl: Control; Statistical significance: *p < 0.05, p < 0.01, *p < 0.001.
The structured workflow and detailed protocols outlined in this application note provide a clear roadmap for bridging the critical validation gap in HCC ncRNA research. By systematically integrating in-silico predictions from bulk and single-cell RNA-seq data with rigorous experimental confirmation, researchers can robustly prioritize and validate ncRNAs with genuine biological and clinical relevance. This approach significantly accelerates the discovery of novel diagnostic biomarkers and therapeutic targets, ultimately contributing to improved outcomes for patients with hepatocellular carcinoma.
Benchmarking and Translating ncRNA Biomarkers into Clinical Potential
In Vitro and In Vivo Functional Validation of Candidate ncRNAs
Hepatocellular carcinoma (HCC) represents a leading cause of cancer-related mortality worldwide, with its pathogenesis involving complex genetic and epigenetic alterations [82] [98]. Next-generation sequencing technologies have revealed that non-coding RNAs (ncRNAs), particularly long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), play crucial regulatory roles in HCC development and progression [82] [99]. These ncRNAs modulate critical cancer-relevant processes including cell cycle regulation, TGF-β signaling, liver metabolism, oxidative phosphorylation, and immune responses [82] [98] [46]. The functional characterization of candidate ncRNAs requires an integrated approach combining robust computational prediction with rigorous experimental validation in both in vitro and in vivo settings [100]. This protocol outlines a standardized framework for validating ncRNA function specifically within HCC research, providing researchers with detailed methodologies for establishing the pathological relevance of ncRNA candidates.
Computational Prediction and Identification of Candidate ncRNAs
Bioinformatics Pipelines for ncRNA Identification
The initial identification of candidate ncRNAs begins with comprehensive RNA sequencing analysis. For HCC tissues, RNA-Seq data should be processed using specialized bioinformatics pipelines designed to handle the unique characteristics of ncRNAs. The Firalink pipeline represents one such tool that can be adapted for HCC studies, providing quality control, contamination screening, alignment, and quantification specifically optimized for ncRNA transcripts [101].
Key steps in the bioinformatics workflow include:
- Quality Control: Assess sequence quality using FastQC to evaluate read counts, duplication levels, Phred quality scores, GC composition, and adapter contamination [101].
- Read Trimming: Remove low-quality sequences and adapters using Trimmomatic or similar tools (Phred score threshold typically set to 30) [101].
- Contamination Screening: Utilize Kraken for taxonomic classification to detect potential bacterial, fungal, or viral contamination in samples [101].
- Alignment and Quantification: Employ pseudo-alignment tools like Kallisto for efficient mapping to ncRNA references, generating count tables for downstream analysis [101].
- Data Compilation: Combine count tables into a unified matrix using Python or R scripts for subsequent differential expression analysis [101].
Differential Expression Analysis and Target Prediction
Following data processing, identify differentially expressed ncRNAs using established statistical frameworks. For HCC studies, compare tumor tissues against paired non-tumorous liver tissues using criteria of |log2FC| ≥ 1 and FDR < 0.05 [82] [102]. Weighted gene co-expression network analysis (WGCNA) can further identify ncRNA modules associated with specific HCC pathological features [82] [46].
For target prediction, utilize integrated databases and tools:
- miRNA Target Prediction: miRWalk, TargetScan, and DIANA-microT-CDS for identifying miRNA-mRNA interactions [100]
- LncRNA Target Prediction: DIANA-LncBase for experimentally supported miRNA targets of lncRNAs [100]
- Interaction Networks: Construct lncRNA-mRNA regulatory networks using Cytoscape to visualize potential interactions [102]
Table 1: Key Bioinformatics Tools for ncRNA Identification and Target Prediction
| Tool Category | Tool Name | Primary Function | Key Features |
|---|---|---|---|
| miRNA Database | miRBase | miRNA sequence repository | 38,589 precursors from 271 organisms [100] |
| miRNA Target Prediction | miRWalk | Target site prediction | Compares results from multiple prediction tools [100] |
| miRNA Target Prediction | TargetScan | Target site prediction | Provides site conservation readouts across species [100] |
| LncRNA Target Prediction | DIANA-LncBase | miRNA-lncRNA interactions | Manually curated interactions from experimental data [100] |
| Network Visualization | Cytoscape | Interaction networks | Constructs lncRNA-mRNA regulatory networks [102] |
| Functional Enrichment | DAVID | Functional annotation | GO and KEGG pathway analysis [102] |
In Vitro Functional Validation of Candidate ncRNAs
Gain-of-Function and Loss-of-Function Approaches
Functional validation of candidate ncRNAs requires modulation of their expression levels in relevant HCC cell lines, followed by assessment of phenotypic outcomes.
Loss-of-Function Strategies:
- CRISPR/Cas9 Knockout: For complete genomic deletion of lncRNA genes, utilize a paired-guide RNA (pgRNA) approach where two sgRNAs flank the promoter region or essential exonic sequences to create large deletions [103]. Design 20+ pgRNAs per target to account for variable deletion efficiency [103].
- CRISPR Interference (CRISPRi): Employ nuclease-deficient Cas9 (dCas9) fused to transcriptional repressor domains (e.g., KRAB) to epigenetically suppress lncRNA transcription without altering genomic DNA [103]. This approach is particularly valuable for lncRNAs where transcription itself may be functional.
- RNA Interference: Utilize siRNA or shRNA for transcript-specific knockdown. This method is effective for both lncRNAs and miRNAs, with careful attention to potential off-target effects.
- Antisense Oligonucleotides (ASOs): Design gapmer ASOs with modified nucleotides (e.g., 2'-O-methoxyethyl) to induce RNase H-mediated degradation of target ncRNAs [103].
Gain-of-Function Strategies:
- Plasmid-Based Overexpression: Clone full-length ncRNA sequences into mammalian expression vectors under strong constitutive promoters (e.g., CMV).
- CRISPR Activation (CRISPRa): Utilize dCas9 fused to transcriptional activators (e.g., VP64-p65-Rta) to enhance endogenous ncRNA expression [103].
- Viral Transduction: Employ lentiviral or adenoviral vectors for stable integration and long-term expression of ncRNA candidates.
Phenotypic Assays for Functional Assessment
Following ncRNA modulation, assess phenotypic changes using standardized assays:
Proliferation and Viability:
- Conduct MTT or CCK-8 assays at 24, 48, and 72 hours post-transfection
- Perform clonogenic assays with 10-14 day incubation to assess long-term proliferation
- Utilize real-time cell analysis (e.g., xCELLigence) for kinetic monitoring
Migration and Invasion:
- Employ Transwell assays with 8μm pore membranes, with Matrigel coating for invasion assays
- Conduct wound healing assays with imaging at 0, 12, 24, and 48 hours
- Use live-cell imaging to track individual cell movements
Apoptosis and Cell Cycle:
- Analyze by flow cytometry using Annexin V/PI staining for apoptosis
- Assess cell cycle distribution with PI/RNase staining
- Examine caspase activation via Western blot or fluorescent assays
Table 2: Key Research Reagent Solutions for In Vitro Functional Validation
| Reagent Category | Specific Examples | Function/Application | Considerations for HCC Research |
|---|---|---|---|
| CRISPR Systems | Cas9, dCas9-KRAB, dCas9-VPR | Genomic editing, transcriptional repression/activation | pgRNA approach recommended for lncRNA knockout [103] |
| RNA Targeting | siRNA, shRNA, ASOs | Transcript knockdown | Gapmer ASOs effective for nuclear lncRNAs [103] |
| Viral Vectors | Lentivirus, Adenovirus | Stable gene delivery | Use for difficult-to-transfect primary hepatocytes |
| Cell Viability Assays | MTT, CCK-8, xCELLigence | Proliferation assessment | Confirm linear range for HCC cell lines used |
| Migration/Invasion Assays | Transwell, Matrigel, Wound Healing | Metastatic potential | Use appropriate ECM components for liver microenvironment |
| Apoptosis Assays | Annexin V/PI, Caspase Glo | Cell death quantification | Establish baseline apoptosis for specific HCC lines |
Molecular Validation of ncRNA Mechanisms
Confirm the mechanistic basis of ncRNA function through molecular analyses:
Expression Validation:
- Extract total RNA using miRNeasy or similar kits with DNase treatment
- Perform reverse transcription with gene-specific primers or random hexamers
- Conduct qRT-PCR using SYBR Green or TaqMan chemistry with appropriate reference genes (e.g., GAPDH, SRSF4) [102] [8]
- Calculate relative expression using the 2^(-ΔΔCt) method with technical triplicates
Subcellular Localization:
- Perform cellular fractionation to separate nuclear and cytoplasmic components
- Validate fraction purity with controls (e.g., U6 for nuclear, GAPDH for cytoplasmic)
- Utilize RNA fluorescence in situ hybridization (FISH) for spatial resolution
Interaction Partner Identification:
- Conduct RNA immunoprecipitation (RIP) to identify protein binding partners
- Perform chromatin isolation by RNA purification (ChIRP) for chromatin-associated lncRNAs
- Implement cross-linking techniques (CLIP) for precise mapping of interaction sites
In Vivo Functional Validation in HCC Models
Animal Models for HCC Research
Select appropriate in vivo models based on research questions and ncRNA conservation:
Xenograft Models:
- Subcutaneous implantation of human HCC cell lines in immunodeficient mice
- Orthotopic implantation into liver parenchyma for more physiologically relevant microenvironment
- Utilize luciferase-tagged cells for longitudinal monitoring of tumor growth
Genetically Engineered Mouse Models:
- Implement hydrodynamic tail vein injection for liver-specific gene delivery
- Use transgenic mice with liver-specific Cre recombinase for conditional gene manipulation
- Consider models with specific HCC driver mutations (e.g., Myc, β-catenin)
Humanized Mouse Models:
- For studying human-specific ncRNAs, employ humanized TK-NOG mice with livers repopulated by human hepatocytes [104]
- This model is particularly valuable for non-conserved human lncRNAs that lack murine counterparts [104]
Experimental Design and Monitoring
Study Timeline and Endpoints:
- Monitor tumor growth weekly via caliper measurements (subcutaneous) or imaging (orthotopic)
- Establish predefined endpoints based on tumor volume (e.g., 1500mm³) or clinical signs
- Collect tissues at sacrifice for molecular and histological analyses
Sample Collection and Processing:
- Harvest tumor tissue, adjacent non-tumor liver, and potential metastatic sites
- Preserve samples in multiple formats: flash-freezing for RNA/protein, formalin-fixation for histology, and optimal cutting temperature compound for frozen sections
- Collect blood for serum biomarker analysis and potential circulating ncRNA detection
In Vivo Delivery Methods for ncRNA Modulation
Viral Vector Delivery:
- Utilize adeno-associated viruses (AAV) with liver-specific promoters (e.g., TBG, ApoE) for sustained ncRNA expression or knockdown
- Implement hydrodynamic injection for high-efficiency, transient delivery to hepatocytes
Oligonucleotide-Based Therapeutics:
- Employ chemically modified ASOs for RNase H-mediated degradation of target ncRNAs [103]
- Utilize lipid nanoparticles for efficient delivery of siRNA or miRNA mimics/inhibitors to liver tissues
- Consider GalNAc-conjugated oligonucleotides for hepatocyte-specific targeting
Integration of HCC-Specific Analytical Approaches
Pathway and Network Analysis in HCC Context
Analyze ncRNA function within established HCC signaling pathways:
Transcriptomic Profiling:
- Conduct RNA-Seq on manipulated HCC cells or tumor tissues to identify differentially expressed genes
- Perform gene set enrichment analysis (GSEA) for pathways relevant to HCC (e.g., Wnt/β-catenin, p53, TGF-β, oxidative phosphorylation) [98] [46]
- Integrate single-cell RNA-Seq data to resolve cellular heterogeneity in HCC tumors [46]
Immune Microenvironment Characterization:
- Utilize CIBERSORT or similar deconvolution algorithms to assess immune cell infiltration from bulk RNA-Seq data [46]
- Perform spatial transcriptomics to map ncRNA expression within tumor immune niches
- Analyze correlation between ncRNA expression and immune checkpoint markers
Biomarker Potential and Clinical Translation
Evaluate the diagnostic and prognostic potential of candidate ncRNAs:
Diagnostic Performance Assessment:
- Quantify ncRNA levels in plasma or serum from HCC patients and controls using qRT-PCR [8]
- Construct receiver operating characteristic (ROC) curves to evaluate sensitivity and specificity
- Compare performance against established biomarkers (e.g., AFP) [8]
Machine Learning Integration:
- Develop predictive models combining multiple ncRNAs with clinical parameters [8]
- Utilize Python's Scikit-learn or similar platforms to build classifiers
- Validate model performance in independent cohorts with appropriate cross-validation
Prognostic Correlation:
- Analyze association between ncRNA expression and clinical outcomes (overall survival, recurrence-free survival)
- Perform multivariate Cox regression to assess independent prognostic value
- Evaluate relationship with treatment response where applicable
Visualization of Experimental Workflows
Integrated ncRNA Validation Pipeline
Functional Screening Strategies for ncRNAs
The functional validation of candidate ncRNAs in HCC requires a multidisciplinary approach integrating computational biology, molecular techniques, and disease-relevant model systems. This protocol provides a comprehensive framework for establishing the pathological significance of ncRNAs, from initial identification through mechanistic characterization and assessment of clinical relevance. As ncRNA research continues to evolve, these standardized methodologies will facilitate the discovery of novel biomarkers and therapeutic targets for hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) represents a significant global health challenge, ranking as the sixth most prevalent cancer worldwide and causing an estimated 750,000 fatalities annually [105]. The disease demonstrates pronounced molecular heterogeneity, with approximately 70% of patients experiencing recurrence within five years following initial treatment [105]. This clinical reality underscores the critical need for robust prognostic models that can stratify patients according to recurrence risk, thereby enabling personalized therapeutic strategies.
Recent advances in high-throughput sequencing technologies have revolutionized HCC prognostic model development. Traditional approaches based solely on clinical parameters are increasingly being supplemented by molecular signatures derived from bulk RNA sequencing, single-cell RNA sequencing (scRNA-seq), and non-coding RNA profiling [105] [57] [106]. The integration of these multi-omics data types with sophisticated statistical learning methods like LASSO Cox regression has enabled the creation of more accurate and biologically informative prognostic tools. These models facilitate risk stratification by identifying key molecular drivers of HCC progression and recurrence, ultimately supporting clinical decision-making for researchers, scientists, and drug development professionals focused on oncology therapeutics.
Theoretical Foundation of LASSO Cox Regression
Principles of Regularization in Survival Analysis
LASSO (Least Absolute Shrinkage and Selection Operator) Cox regression represents a powerful regularization technique that addresses the critical challenge of high-dimensional data in prognostic modeling, where the number of potential predictors (p) often far exceeds the number of observations (n) [107]. This method operates by imposing an L1-norm penalty on the regression coefficients, effectively shrinking less important coefficients toward zero and performing automatic variable selection simultaneously [108]. The fundamental strength of LASSO lies in its ability to generate sparse models with only a subset of non-zero coefficients, thereby enhancing model interpretability – a crucial consideration for clinical applications [108].
The mathematical formulation of the LASSO Cox regression optimizes the following objective function:
$$\text{argmax}{\beta} \left\{ \ell(\beta) - \lambda \sum{j=1}^{p} |\beta_j| \right\}$$
where $\ell(\beta)$ represents the partial log-likelihood of the Cox model, $\beta_j$ denotes the regression coefficients, and $\lambda$ is the tuning parameter that controls the strength of the penalty term. Through this mechanism, LASSO effectively balances model complexity with predictive accuracy, preventing overfitting while maintaining essential prognostic signals [108] [107].
Comparative Advantages in HCC Research
In the context of HCC research, LASSO Cox regression offers distinct advantages over traditional statistical methods. Conventional Cox proportional hazards models become unstable or infeasible when dealing with high-dimensional genomic data, as the number of candidate genes or non-coding RNAs frequently numbers in the thousands while patient cohorts typically comprise only hundreds of individuals [109] [107]. LASSO addresses this limitation by selecting the most informative molecular features from large candidate pools, making it particularly suited for identifying multi-gene signatures from transcriptomic data [105] [110].
Furthermore, LASSO demonstrates superior performance in feature selection compared to Ridge regression (which uses L2-norm penalty) in scenarios where the underlying true model is sparse – an assumption that generally holds for molecular prognostic markers, where only a small subset of transcripts typically carries significant predictive information [108]. This property makes LASSO ideal for developing parsimonious prognostic models that integrate seamlessly with clinical workflows, requiring measurement of only a limited number of biomarkers for practical implementation.
Integrated Analysis of Single-Cell and Bulk RNA Sequencing Data
Experimental Workflow for Data Integration
The integration of scRNA-seq and bulk RNA-seq data represents a cutting-edge approach for identifying robust prognostic signatures in HCC. This methodology leverages the complementary strengths of both technologies: scRNA-seq provides unprecedented resolution of cellular heterogeneity within tumors, while bulk RNA-seq offers clinical correlative power through larger sample sizes and associated outcome data [105] [57]. The experimental workflow encompasses multiple stages, from data acquisition through model validation, as illustrated below:
Protocol for Integrated scRNA-seq and Bulk RNA-seq Analysis
Data Acquisition and Quality Control
- Data Sources: Obtain scRNA-seq datasets from GEO (e.g., GSE242889 with 5 HCC patients) and bulk RNA-seq data from TCGA-LIHC (342 patients) and GEO (e.g., GSE76427 with 108 patients) [105]
- Quality Control for scRNA-seq: Filter cells with <200 detected genes or >4,000 genes (potential multiplets), and exclude cells with mitochondrial transcript proportions ≥50% using Seurat package [105]
- Quality Control for Bulk RNA-seq: Retain samples with complete recurrence-free survival (RFS) data and RNA-seq profiles, excluding cases with missing clinical annotations [105]
Data Processing and Integration
- scRNA-seq Processing: Normalize data using SCTransform, perform dimensionality reduction via PCA, and cluster cells using FindClusters algorithm in Seurat (resolution parameter = 0.8) [105] [57]
- Cell Type Annotation: Identify hepatocyte populations using canonical marker genes and extract expression data for subsequent analysis [105]
- Differential Expression Analysis: Identify differentially expressed genes (DEGs) using FindMarkers in Seurat for scRNA-seq (|log2FC|>0.5, adjusted p<0.05) and limma R package for bulk RNA-seq (|log2FC|>1, adjusted p<0.05) [105]
- Data Integration: Cross-reference DEGs from both platforms to identify consensus candidates, then evaluate their association with RFS using survival analysis (log-rank p<0.05) [105]
Feature Selection and Model Construction
- Candidate Gene Pool: Begin with survival-associated DEGs common to both sequencing platforms (e.g., 53 genes identified in recent study) [105]
- LASSO Cox Regression: Implement using glmnet R package (version 4.1) with ten-fold cross-validation to determine optimal lambda value [105]
- Gene Signature Development: Construct prognostic model based on genes with non-zero coefficients in the final LASSO model [105]
Table 1: Exemplar Multi-Gene Signatures from Integrated Analysis in HCC
| Study Reference | Gene Signature | Number of Genes | Predictive Performance (AUC) | Clinical Endpoint |
|---|---|---|---|---|
| Zhou et al. [105] | CDKN2A, CFHR3, CYP2C9, HMGB2, IGLC2, JPT1 | 6 | Validated in independent cohort | Recurrence-free survival |
| PANoptosis Study [106] | CYBC1, JPT1, UQCRH, YIF1B | 4 | Time-dependent AUC for 1/3/5-year survival | Overall survival |
| Immune Model [110] | Immune-related gene signature | 6 | 0.85, 0.779, 0.857 for 1/3/5-year survival | Overall survival |
Technical Considerations and Optimization
The integrated analysis approach requires careful consideration of several technical factors. Batch effects between different datasets must be addressed using harmonization algorithms such as Harmony [105]. For scRNA-seq data, the choice of clustering resolution significantly impacts cell type identification and subsequent DEG analysis [105]. Additionally, the threshold for defining DEGs should be optimized based on data quality and sample size, with more stringent criteria applied to bulk RNA-seq data due to its lower biological noise compared to scRNA-seq [105].
The computational intensity of integrated analysis necessitates appropriate infrastructure, particularly for processing large scRNA-seq datasets containing >50,000 cells [57]. Implementation in R using Seurat for scRNA-seq analysis and glmnet for LASSO regression provides a robust, reproducible framework. Finally, biological validation of identified signatures through experimental approaches such as RT-qPCR on patient tissues remains essential to confirm clinical utility [105].
Development of Non-Coding RNA Prognostic Signatures
Long Non-Coding RNA Profiling and Analysis
Long non-coding RNAs (lncRNAs) have emerged as crucial regulators of oncogenesis and progression in HCC, offering substantial potential as prognostic biomarkers due to their tissue-specific expression patterns and functional diversity [109] [8]. The protocol for developing lncRNA-based prognostic signatures encompasses distinct phases from discovery through clinical application, as detailed below:
Protocol for lncRNA-Based Signature Development
Cohort Selection and Stratification
- Patient Inclusion: Select HCC patients with histologically confirmed diagnosis, complete RNA-Seq data for lncRNAs, available fibrosis status (Ishak score), and survival outcomes [109]
- Fibrosis Stratification: Categorize patients as "with fibrosis" (portal fibrosis, fibrous septum, nodular formation, incomplete cirrhosis, established cirrhosis) or "without fibrosis" (Ishak score: no fibrosis) [109]
- Sample Size Considerations: Ensure adequate statistical power by including sufficient patients in each stratum (e.g., 135 with fibrosis, 72 without fibrosis in published study) [109]
lncRNA Expression Profiling
- Data Acquisition: Obtain lncRNA expression profiles from TCGA via UCSC Xena or perform original sequencing using RNA extraction from HCC tissues [109]
- Quality Control: Filter lncRNAs with expression levels of 0 in >50% of patients using edgeR algorithm in R [109]
- Differential Expression: Identify DElncRNAs between HCC patients with/without fibrosis using |log2FC|>1 with FDR<0.05 as thresholds [109]
Signature Construction and Validation
- Univariate Cox Analysis: Test association between DElncRNAs and overall survival (OS) or recurrence-free survival (RFS) with p<0.05 significance [109]
- LASSO Cox Regression: Apply glmnet package to identify optimal lncRNA combination, using ten-fold cross-validation to prevent overfitting [109] [111]
- Risk Score Calculation: Compute individual risk scores using formula: Risk score = (β1 × expression lncRNA1) + (β2 × expression lncRNA2) + ... + (βn × expression lncRNAn) [109]
- Validation: Assess prognostic performance using time-dependent ROC curves and validate in independent cohorts [109]
Table 2: Exemplar lncRNA-Based Prognostic Signatures in HCC
| Patient Subgroup | Signature Purpose | Number of lncRNAs | Representative lncRNAs | Performance (AUC) |
|---|---|---|---|---|
| With Fibrosis [109] | OS Prediction | 5 | AL359853.1, Z93930.3, HOXA-AS3 | >0.7 |
| With Fibrosis [109] | RFS Prediction | 12 | PLCE1-AS1, Z93930.3, LINC02273 | >0.7 |
| Without Fibrosis [109] | OS Prediction | 7 | LINC00239, HOXA-AS3, NRIR | >0.7 |
| Without Fibrosis [109] | RFS Prediction | 5 | AC021744.1, NRIR, LINC00487 | >0.7 |
| Cuproptosis-Related [111] | OS Prediction | 3 | PICSAR, FOXD2-AS1, AP001065.1 | 0.741 |
| Plasma-Based [8] | Diagnosis & Prognosis | 4 | LINC00152, LINC00853, UCA1, GAS5 | 100% sensitivity, 97% specificity |
Advanced lncRNA Signature Methodologies
Recent methodological innovations have enhanced the sophistication of lncRNA-based prognostic models. Machine learning integration with lncRNA expression data has demonstrated remarkable diagnostic performance, achieving up to 100% sensitivity and 97% specificity when combining lncRNA profiles with conventional laboratory parameters [8]. The ratio-based approaches, such as the LINC00152 to GAS5 expression ratio, have shown significant correlation with mortality risk, providing simplified metrics for clinical implementation [8].
For functional annotation, co-expression analysis with mRNAs can illuminate the biological processes underpinning lncRNA signatures, with common enriched pathways including cell cycle regulation, chemokine signaling, Th17 cell differentiation, and thermogenesis [109]. Furthermore, liquid biopsy applications using plasma-circulating lncRNAs offer non-invasive alternatives for dynamic monitoring of disease progression and treatment response [8].
The incorporation of mechanistic themes such as cuproptosis – a novel form of copper-dependent programmed cell death – has enabled the development of biologically grounded signatures with enhanced prognostic capability [111]. These cuproptosis-related lncRNA models not only predict survival but also inform about immune infiltration patterns and potential response to immunotherapy, creating opportunities for personalized treatment approaches [111].
LASSO-Cox Regression Protocol for HCC Prognostic Modeling
Step-by-Step Computational Implementation
The practical implementation of LASSO Cox regression for HCC prognostic modeling requires meticulous execution of sequential computational steps:
Data Preparation and Preprocessing
- Expression Data Processing: Normalize raw count data from RNA-seq using VST transformation (bulk) or SCTransform (single-cell) and log2-transform for analysis [105] [57]
- Survival Data Integration: Merge expression matrices with corresponding clinical data, ensuring accurate time-to-event information for recurrence-free survival (RFS) or overall survival (OS) [105]
- Data Splitting: Partition dataset into training (70%) and validation (30%) sets using methods like Kennard-Stone algorithm to ensure representative distribution [107]
Feature Pre-selection and Dimension Reduction
- Univariate Screening: Perform initial univariate Cox regression on all candidate genes to identify those with significant survival association (p<0.05) [109]
- Variance Filtering: Remove low-variance genes (bottom 20%) to reduce noise and computational burden [57]
- Collinearity Check: Assess correlation between top candidates and remove highly correlated features (r>0.8) to minimize redundancy [110]
LASSO Cox Regression Implementation
- Package Implementation: Utilize glmnet R package (version 4.1) with family="cox" parameter setting [105]
- Lambda Selection: Perform k-fold cross-validation (typically k=10) to identify optimal lambda value – typically lambda.min or lambda.1se for more parsimonious models [105] [109]
- Model Fitting: Execute LASSO Cox regression with optimized lambda to obtain regression coefficients for selected features [105]
Risk Score Calculation and Stratification
- Score Computation: Calculate risk score for each patient using formula: Risk score = Σ(βi × Expi), where βi represents coefficients and Expi denotes expression levels [105]
- Cut-off Determination: Dichotomize patients into high- and low-risk groups using median risk score or optimal cut-point determined by maximally selected rank statistics [105] [109]
- Stratification Validation: Confirm stratification efficacy through Kaplan-Meier analysis with log-rank test (p<0.05 indicating significant separation) [105]
Model Performance Assessment
- Discrimination Ability: Evaluate using time-dependent receiver operating characteristic (ROC) curves and calculate area under curve (AUC) for 1-, 3-, and 5-year survival [109] [110]
- Calibration: Assess agreement between predicted and observed outcomes using calibration plots [110]
- Clinical Utility: Perform decision curve analysis (DCA) to evaluate net benefit across different threshold probabilities [107]
Advanced Model Development and Validation
Multi-Omics Model Integration
- Combined Signatures: Integrate gene expression signatures with clinical parameters (BCLC stage, tumor size, liver function) using multivariate Cox regression [108]
- Nomogram Construction: Develop comprehensive prognostic nomograms that combine molecular risk scores with clinical variables for enhanced prediction [110] [107]
- Validation Cohorts: Confirm model performance in independent datasets (e.g., TCGA training with GEO validation) to ensure generalizability [105]
Biological Interpretation and Functional Analysis
- Pathway Enrichment: Perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on signature genes using clusterProfiler package [105] [57]
- Immune Microenvironment: Evaluate immune cell infiltration patterns using CIBERSORT or ssGSEA and correlate with risk scores [110]
- Therapeutic Implications: Explore associations between risk strata and drug sensitivity using pRRophetic package or similar tools [106]
Research Reagent Solutions
Table 3: Essential Research Reagents for HCC Prognostic Model Development
| Reagent/Resource | Specification | Application | Example Sources |
|---|---|---|---|
| RNA Extraction | miRNeasy Mini Kit | High-quality RNA from tissues/cells | QIAGEN (cat no. 217004) [8] |
| cDNA Synthesis | RevertAid First Strand cDNA Synthesis Kit | cDNA preparation for qPCR | Thermo Scientific (cat no. K1622) [8] |
| qRT-PCR | PowerTrack SYBR Green Master Mix | lncRNA expression quantification | Applied Biosystems (cat no. A46012) [8] |
| scRNA-seq Platform | 10X Genomics with Seurat v4.3.0 | Single-cell transcriptome profiling | GSE202642, GSE149614 [57] [106] |
| Cell Culture | DMEM with 10% FBS, 1% penicillin/streptomycin | Maintenance of HCC cell lines | Gibco [106] |
| Antibodies | Anti-YIF1B, anti-GAPDH | Protein validation via Western blot | Abcam (ab188127), Proteintech (10494-1-AP) [106] |
| Flow Cytometry | Anti-CD45, anti-CD8, anti-NK1.1 | Immune cell phenotyping | BioLegend (103130, 563786, 108708) [106] |
| LASSO Implementation | glmnet R package (v4.1) | Regularized Cox regression | CRAN repository [105] |
| Survival Analysis | survival R package (v3.2.7) | Survival modeling and validation | CRAN repository [105] |
The integration of LASSO Cox regression with multi-omics data represents a transformative approach for developing robust prognostic models in hepatocellular carcinoma. This methodology effectively addresses the high-dimensionality challenge inherent in transcriptomic data while generating interpretable models with direct clinical relevance. The protocols outlined herein provide a comprehensive framework for constructing prognostic signatures through integrated analysis of single-cell and bulk RNA sequencing data, with particular emphasis on non-coding RNA biomarkers.
Future developments in this field will likely focus on multi-modal integration of genomic, transcriptomic, epigenomic, and proteomic data to capture the full complexity of HCC pathogenesis. Additionally, the incorporation of digital pathology features and radiomic profiles may further enhance predictive accuracy. As single-cell technologies advance, spatial transcriptomics will enable the interrogation of gene expression within architectural context, providing unprecedented insights into tumor microenvironment interactions. The continued refinement of these computational approaches will accelerate the development of personalized prognostic tools that ultimately improve clinical decision-making and patient outcomes in hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) is the most common form of primary liver cancer and a leading cause of cancer-related mortality worldwide. The limitations of traditional biomarkers like Alpha-fetoprotein (AFP), which lacks sufficient sensitivity and specificity for early detection, have driven the exploration of novel molecular signatures. Within this context, RNA sequencing analyses have revealed that non-coding RNAs (ncRNAs) are critically involved in HCC tumorigenesis, progression, and metastasis. This Application Note provides a detailed comparative analysis and structured protocols for evaluating the performance of emerging ncRNA signatures against traditional AFP in HCC management.
Performance Comparison: ncRNA Signatures vs. Traditional Markers
Table 1: Diagnostic Performance of ncRNA Signatures vs. Traditional Serum Markers
| Biomarker Category | Specific Marker / Signature | Sensitivity Range | Specificity Range | AUC / Key Performance Metric | Key Advantages |
|---|---|---|---|---|---|
| Traditional Serum Marker | AFP (>20 ng/mL) | ~60% [112] [8] | -- | -- | Well-established, low cost, widely available [69] |
| AFP (>400 ng/mL) | ~30-40% of HCCs are AFP-negative (<20 ng/mL) [113] | -- | -- | Diagnostic criterion at high levels [8] | |
| Single lncRNA | LINC00152 | 83% [8] | 67% [8] | -- | Detected in plasma, suitable for liquid biopsy [8] |
| UCA1 | 81% [8] | 53% [8] | -- | Detected in plasma, suitable for liquid biopsy [8] | |
| GAS5 | 60% [8] | 67% [8] | -- | Tumor suppressor function [8] | |
| LINC00853 | 63% [8] | 67% [8] | -- | Detected in plasma, suitable for liquid biopsy [8] | |
| Multi-lncRNA Signature | 3-DRL Signature (AC016717.2, AC124798.1, AL031985.3) | -- | -- | 1-yr AUC: 0.756; 3-yr AUC: 0.695; 5-yr AUC: 0.701 [114] | Predicts overall survival, associates with immune function and drug sensitivity [114] |
| 4-lncRNA Panel (LINC00152, LINC00853, UCA1, GAS5) + Machine Learning | 100% [8] | 97% [8] | -- | Superior to individual lncRNAs or AFP alone [8] | |
| Composite Clinical Score | GALAD Score | 73% (Early-Stage HCC) [69] | 87% (Early-Stage HCC) [69] | AUROC: 0.92 [69] | Integrates gender, age, AFP, AFP-L3, and DCP [69] |
Table 2: Prognostic and Therapeutic Utility of ncRNA Signatures vs. AFP
| Characteristic | Traditional AFP | ncRNA Signatures |
|---|---|---|
| Prognostic Value | Limited independent prognostic value within normal range (<20 ng/mL) [115]. High-normal levels (7-20 ng/mL) linked to poorer liver function and more tumors, but not an independent prognostic factor in multivariate analysis [115]. | Powerful prognostic value. A 3-Disulfidptosis-Related lncRNA (DRL) signature effectively stratifies patients into high-risk and low-risk groups with significantly different overall survival (p<0.001) [114]. |
| Therapeutic Implications | Level does not directly inform therapy selection. | Signatures can predict drug sensitivity. The 3-DRL signature shows significant differences in drug sensitivity between high-risk and low-risk groups, potentially guiding personalized therapy [114]. |
| Insight into Biology | Reflects hepatocyte differentiation but mechanistic role in HCC is not fully defined. | Provide direct mechanistic insight. They regulate key pathways: proliferation (e.g., HULC, NEAT1), metastasis (e.g., HOTAIR), apoptosis (e.g., GAS5), and metabolism (e.g., linc-RoR in hypoxia) [27] [28]. |
Application Notes & Experimental Protocols
Protocol 1: Developing a Prognostic ncRNA Signature from RNA-Seq Data
This protocol outlines the process for identifying and validating a prognostic long non-coding RNA (lncRNA) signature for Hepatocellular Carcinoma (HCC), based on the methodology used in disulfidptosis-related research [114].
Workflow Diagram: Prognostic ncRNA Signature Development
Procedure
Data Acquisition and Preprocessing:
- Source: Obtain HCC transcriptomic data (RNA-Seq) and corresponding clinical data (survival, stage, etc.) from public repositories like The Cancer Genome Atlas (TCGA) (e.g., 422 samples: 373 tumor, 49 normal) [114] [116] or Gene Expression Omnibus (GEO).
- Processing: Normalize raw count data (e.g., using DESeq2 or edgeR) and annotate lncRNAs based on a reference genome (e.g., GENCODE) [112].
Identification of Phenotype-Associated lncRNAs:
- Define Target Phenotype: Select a specific biological process or form of cell death (e.g., disulfidptosis, apoptosis, immune response). Compile a list of known related genes (e.g., 22 disulfidptosis-related genes) from literature [114].
- Correlation Analysis: Perform Spearman correlation analysis between the phenotype-related genes and all expressed lncRNAs. Filter for significant correlations (e.g., |R| > 0.5, P < 0.001) to identify a candidate set of phenotype-related lncRNAs (DRLs) [114].
Signature Construction and Validation:
- Prognostic Screening: Conduct univariate Cox regression analysis on the candidate lncRNAs to identify those significantly associated with overall survival (P < 0.01).
- Variable Selection: Apply the Least Absolute Shrinkage and Selection Operator (LASSO) Cox regression to the significant lncRNAs from the previous step to prevent overfitting and select the most robust predictors for the final model [114].
- Model Building: Use multivariate Cox proportional hazards regression on the LASSO-selected lncRNAs to build the final prognostic model. The risk score for each patient is calculated as a linear combination of the expression levels of the final lncRNAs weighted by their regression coefficients. Example:
Risk Score = (exp lncRNA1 * coef1) + (exp lncRNA2 * coef2) + ...[114]. - Cohort Assignment: Split the patient cohort into training and validation sets (e.g., 1:1 ratio). Calculate the median risk score in the training set and use it to classify all patients into high-risk and low-risk groups.
- Validation: Validate the signature's performance in the independent validation set using Kaplan-Meier survival analysis (log-rank test) and time-dependent Receiver Operating Characteristic (ROC) analysis (e.g., for 1, 3, and 5-year survival) [114].
Downstream Functional Analysis:
- Pathway Enrichment: Use the "limma" R package to identify differentially expressed genes (DEGs) between high-risk and low-risk groups. Perform functional enrichment analysis (Gene Ontology - GO, and Kyoto Encyclopedia of Genes and Genomes - KEGG) on the DEGs with the "clusterProfiler" R package to understand the underlying biological pathways [114].
- Immune and Therapeutic Profiling: Evaluate differences in the tumor immune microenvironment (e.g., immune cell infiltration using CIBERSORT), tumor mutational burden (TMB), and drug sensitivity (e.g., using GDSC database and "oncoPredict" R package) between the risk groups [114].
Protocol 2: Validation of Circulating lncRNAs as Liquid Biopsy Biomarkers
This protocol details the steps for quantifying plasma lncRNAs and developing a diagnostic model, integrating methods from recent studies [112] [8].
Workflow Diagram: Circulating lncRNA Validation and Model Integration
Procedure
Cohort Establishment and Sample Collection:
- Participants: Recruit newly diagnosed, treatment-naïve HCC patients (diagnosed per LI-RADS or histology) and age-matched control groups (healthy individuals, patients with chronic hepatitis B/C, or cirrhosis) [8].
- Ethics: Obtain written informed consent and institutional review board (IRB) approval.
- Sample Collection: Collect peripheral blood in EDTA tubes. Process plasma by centrifuging whole blood at a minimum of 2000 x g for 10 minutes to remove cells and platelets. Aliquot and store plasma at -80°C.
RNA Isolation from Plasma:
- Use a commercial kit designed for liquid biopsies, such as the miRNeasy Mini Kit (QIAGEN), to extract total RNA from plasma samples according to the manufacturer's protocol [8]. Include a spike-in synthetic RNA for normalization if required.
cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR):
- Reverse Transcription: Synthesize cDNA from the extracted RNA using a Reverse Transcription Kit (e.g., RevertAid First Strand cDNA Synthesis Kit, Thermo Scientific) [8].
- qRT-PCR: Perform qRT-PCR reactions in triplicate using a SYBR Green Master Mix on a real-time PCR system (e.g., ViiA 7, Applied Biosystems). Use GAPDH or other stable non-coding RNAs as a housekeeping gene for normalization.
- Primers: Use validated, sequence-specific primers for the target lncRNAs (e.g., LINC00152, UCA1). Calculate relative expression using the 2^(-ΔΔCt) method [8].
Statistical Analysis and Machine Learning Model Integration:
- Individual Performance: Assess the diagnostic power of each lncRNA and AFP alone by generating ROC curves and calculating AUC, sensitivity, and specificity.
- Model Building: Integrate the qRT-PCR data (lncRNA expression levels) with standard clinical laboratory parameters (e.g., AFP, ALT, AST, bilirubin) using a machine learning platform like Python's Scikit-learn.
- Model Training and Evaluation: Train a classifier (e.g., logistic regression, random forest) and evaluate its performance using cross-validation. The model achieving 100% sensitivity and 97% specificity for HCC diagnosis, as demonstrated in prior research, significantly outperforms single biomarkers [8].
Table 3: Essential Reagents and Resources for HCC ncRNA Research
| Item | Function / Application | Example Product / Source |
|---|---|---|
| Total RNA Extraction Kit | Isolation of high-quality RNA (including small RNAs) from tissues or liquid biopsies. | miRNeasy Mini Kit (QIAGEN) [8] |
| Reverse Transcription Kit | Synthesis of complementary DNA (cDNA) from RNA templates for subsequent PCR. | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) [8] |
| qRT-PCR Master Mix | Sensitive and specific quantification of target lncRNA transcripts. | PowerTrack SYBR Green Master Mix (Applied Biosystems) [8] |
| Public Transcriptomic Data | Source of RNA-seq data for discovery-phase analysis and validation. | The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) [114] [46] |
| Bioinformatic Tools | R packages for differential expression, survival, and enrichment analysis. | "limma", "survival", "clusterProfiler" R packages [114] |
| Immune Deconvolution Algorithm | Computational assessment of immune cell infiltration from bulk RNA-seq data. | CIBERSORT [114] [46] |
| Drug Sensitivity Database | Resource for predicting chemotherapeutic response based on genomic features. | Genomics of Drug Sensitivity in Cancer (GDSC) [114] |
Hepatocellular carcinoma (HCC) represents a significant global health challenge, characterized by poor prognosis and high mortality rates, particularly when diagnosis is delayed. [28] The complex molecular landscape of HCC, driven by factors such as chronic hepatitis B virus (HBV) infection, necessitates advanced diagnostic approaches. [117] Within this context, machine learning (ML) has emerged as a transformative technology, capable of identifying complex patterns within clinical, imaging, and molecular data to enable earlier and more accurate HCC detection. [72] Simultaneously, research into the RNA sequencing analysis of non-coding RNAs (ncRNAs) has revealed their critical roles as regulatory molecules in HCC pathogenesis, offering a rich source of potential biomarkers. [28] [118] This document explores the integration of these two frontiers, evaluating the performance of ML models for HCC diagnosis and their synergistic potential with ncRNA research to advance clinical practice and therapeutic development.
Performance of Machine Learning Models in HCC Diagnosis
Machine learning models have demonstrated exceptional performance in diagnosing HCC using various data modalities, from routine clinical variables to advanced radiomic features. The following table summarizes the reported performance metrics of several state-of-the-art models.
Table 1: Performance Metrics of Machine Learning Models for HCC Diagnosis
| Model Type | Data Modality | Cohort Description | Key Performance Metrics | Top Features Identified |
|---|---|---|---|---|
| Random Forest [117] | Clinical & Biochemical | 1,051 HBV-related cACLD patients | AUC: 0.979, Accuracy: 0.977, Sensitivity: 0.808 | LSM, Age, Platelet, Bile Acid, WBC |
| Random Forest [72] | Clinical & Serologic | Filipino cohort (73 HCC, 658 non-HCC) | AUC: 0.999, Accuracy: 98.9%, Sensitivity: 90.5% | AFP, DCP, Age, ALP, AST, Albumin, Platelet |
| LightGBM [72] | Clinical & Serologic | Filipino cohort (73 HCC, 658 non-HCC) | AUC: 0.999, Accuracy: 99.1%, Sensitivity: 94.9% | AFP, DCP, Age, ALP, AST, Albumin, Platelet |
| Radiomics (Combined Model) [119] | Multi-sequence MRI | 321 patients from multiple centers | Accuracy: 0.829 (for predicting pathological grade) | Features from AP, T2WI, and DWI sequences |
| HTRecNet (Deep Learning) [120] | Histopathological Images | 5,432 images (Normal, HCC, CCA) | AUC > 0.99, Accuracy: 0.97 (external test) | Automated feature extraction from tissue images |
The high performance of models like Random Forest and LightGBM, even with a minimal set of 7 clinical predictors, underscores the power of ML to extract profound insights from standardized, accessible data. [72] This is particularly advantageous for resource-limited settings. Furthermore, the application of ML extends beyond mere detection to grading tumors non-invasively via radiomics. [119] The high accuracy of the HTRecNet model in distinguishing HCC from cholangiocarcinoma (CCA) and normal tissue on histopathology images also highlights the potential for ML to augment pathological diagnosis. [120]
Connecting HCC ncRNA Research with Diagnostic Models
The molecular landscape of HCC is profoundly influenced by non-coding RNAs (ncRNAs), which include long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs). These molecules are pivotal regulators of gene expression and play key roles in hepatocarcinogenesis, making them attractive subjects for diagnostic and therapeutic development. [28] [118]
Table 2: Key Non-Coding RNAs in Hepatocellular Carcinoma
| ncRNA Type | Example | Expression in HCC | Proposed Mechanism of Action | Clinical Relevance |
|---|---|---|---|---|
| Oncogenic miRNA | miR-21 [118] | Overexpressed in 82% of tissues | Targets tumor suppressor PTEN, activating PI3K/AKT signaling | Serum sensitivity 78% for diagnosis |
| Tumor Suppressive miRNA | miR-122 [118] | Downregulated in 65% of cases | Represses oncogenes like c-Myc; enhances sorafenib sensitivity | Low expression predicts poor OS (16 vs. 28 months) |
| Oncogenic lncRNA | HOTAIR [28] [118] | Overexpressed in advanced HCC | Promotes chromatin remodeling via interaction with PRC2 | High expression linked to 3-fold higher recurrence rate |
| Oncogenic lncRNA | MALAT1 [118] | Elevated in sorafenib-resistant cells | Sponges miR-143, releasing SNAIL to drive drug resistance | Associated with therapy resistance |
| Oncogenic circRNA | CDR1as [118] | Upregulated 3.5-fold | Sponges miR-7 to activate EGFR signaling | Correlates with vascular invasion (OR=2.3) |
The integration of ncRNA biomarkers into ML diagnostic models represents a promising frontier. For instance, a panel of three miRNAs (miR-21, miR-155, miR-122) has been shown to achieve an AUC of 0.89 for distinguishing HCC from cirrhosis, outperforming the traditional biomarker AFP (AUC=0.72). [118] These ncRNA signatures can provide a quantitative, molecular-level input that could significantly enhance the accuracy and biological interpretability of ML models, moving beyond purely clinical and radiological parameters.
Experimental Protocols
Protocol 1: Developing a Clinical-Based ML Model for HCC Detection
This protocol outlines the steps for building a machine learning model similar to the one described in the Filipino cohort study. [72]
I. Data Collection and Preprocessing
- Patient Cohort: Retrospectively enroll confirmed HCC patients and control subjects (e.g., patients with chronic liver disease without HCC). Ethical approval is mandatory.
- Clinical Variables: Collect a comprehensive set of baseline data, including:
- Demographics: Age, sex.
- Liver Function: Albumin, Alkaline Phosphatase (ALP), Aspartate Transaminase (AST), Alanine Aminotransferase (ALT), Total Bilirubin.
- Tumor Markers: Alpha-fetoprotein (AFP), Des-gamma-carboxy prothrombin (DCP).
- Complete Blood Count: Platelet count, White Blood Cell count.
- Coagulation: International Normalized Ratio (INR).
- Data Cleansing: Handle missing values (e.g., using imputation or deletion) and normalize continuous variables to a standard scale (e.g., Z-score normalization).
II. Feature Selection and Model Training
- Feature Selection: Apply multiple feature selection techniques to identify the most predictive variables.
- Data Splitting: Randomly split the dataset into a training cohort (e.g., 70-80%) and a hold-out validation cohort (e.g., 20-30%).
- Model Building:
III. Model Validation and Interpretation
- Performance Evaluation: Test the trained models on the held-out validation set. Calculate key metrics: Area Under the Curve (AUC), Accuracy, Sensitivity, Specificity, Positive Predictive Value (PPV), and Negative Predictive Value (NPV). [72]
- Model Interpretation: Employ SHapley Additive exPlanations (SHAP) analysis to interpret the model's predictions and understand the contribution and direction of each feature. [117] [119]
Protocol 2: RNA Sequencing Analysis of ncRNAs from HCC Tissues
This protocol provides a framework for generating ncRNA expression data, which can be integrated into ML models as molecular features.
I. Sample Collection and RNA Extraction
- Tissue Samples: Obtain paired HCC and adjacent non-tumor liver tissues from patients undergoing surgical resection, with informed consent and IRB approval. Snap-freeze tissues in liquid nitrogen and store at -80°C.
- RNA Extraction: Homogenize tissue samples and perform total RNA extraction using a TRIzol-based method or a commercial kit. Treat samples with DNase I to remove genomic DNA contamination. [11]
- Quality Control: Assess RNA integrity and purity using an Agilent Bioanalyzer or similar system. Only samples with an RNA Integrity Number (RIN) >7.0 should be used for sequencing.
II. Library Preparation and Sequencing
- ncRNA Enrichment: For miRNA sequencing, use size selection to enrich for small RNAs. For lncRNA and circRNA sequencing, deplete ribosomal RNA (rRNA) from the total RNA.
- Library Construction: Prepare sequencing libraries using kits specific for the ncRNA type of interest (e.g., small RNA library prep kit for miRNAs). Include unique molecular identifiers (UMIs) to correct for PCR duplicates.
- Sequencing: Perform high-throughput sequencing on an Illumina platform (e.g., NovaSeq) to generate a minimum of 50 million paired-end reads per sample.
III. Bioinformatic Analysis and Integration
- Data Processing:
- Quality Control & Trimming: Use FastQC for quality assessment and Trimmomatic to remove adapter sequences and low-quality bases.
- Alignment: Map reads to the human reference genome (e.g., GRCh38) using a splice-aware aligner like STAR.
- Quantification: For lncRNAs and miRNAs, use featureCounts to generate expression counts. For circRNAs, use specialized tools like CIRI2 or find_circ to identify back-splice junctions and quantify expression.
- Differential Expression: Identify significantly dysregulated ncRNAs between HCC and normal tissues using R packages like DESeq2 or edgeR. A false discovery rate (FDR) < 0.05 and |log2 fold change| > 1 are common thresholds.
- Validation: Validate key findings using quantitative RT-PCR (qRT-PCR) on an independent set of samples. [11]
Table 3: Essential Reagents and Tools for HCC ML and ncRNA Research
| Category | Item | Function/Application | Example/Specification |
|---|---|---|---|
| Clinical Data | Liver Stiffness Measurement (LSM) | Key non-invasive predictor of fibrosis and HCC risk. [117] | Transient Elastography (e.g., FibroScan) |
| Serum Biomarkers | Alpha-fetoprotein (AFP), DCP | Critical input features for clinical ML models. [72] | Electrochemiluminescence Immunoassay (ECLIA) |
| RNA Extraction | TRIzol Reagent | For high-quality total RNA isolation from tissues and cells. [11] | Phenol and guanidine isothiocyanate-based solution |
| Sequencing | rRNA Depletion Kit; Small RNA Library Prep Kit | Preparation of libraries for lncRNA/circRNA and miRNA sequencing. | Kits from Illumina, NEB, or Thermo Fisher |
| Computational Tools | SHAP (SHapley Additive exPlanations) | Interpreting ML model output and feature importance. [117] [119] | Python library |
| Bioinformatics Software | DESeq2 / edgeR | Identifying differentially expressed ncRNAs from RNA-seq data. | R/Bioconductor packages |
| Validation | SYBR Green qPCR Master Mix | Quantifying expression levels of candidate ncRNAs. [11] | Includes DNA-binding dye and hot-start polymerase |
Machine learning models have demonstrated remarkable accuracy in diagnosing hepatocellular carcinoma, leveraging diverse data types from clinical variables to radiomic features. Their ability to utilize minimal, cost-effective predictor sets makes them particularly promising for improving diagnostics across varied healthcare settings. The concurrent advancement in understanding the roles of non-coding RNAs in HCC provides a powerful molecular framework. The future of HCC diagnosis and prognosis lies in the integration of these two fields. Developing multi-modal ML models that incorporate both robust clinical data and biologically significant ncRNA biomarkers will likely yield the next generation of highly accurate, interpretable, and clinically actionable tools for managing this complex disease.
The aggressive nature of hepatocellular carcinoma (HCC) and its frequent late-stage diagnosis significantly contribute to poor patient outcomes, with traditional biomarkers like alpha-fetoprotein (AFP) demonstrating limited sensitivity and specificity, particularly in early-stage disease [70]. Liquid biopsy, which enables the isolation and analysis of tumor-derived components from bodily fluids, presents a promising non-invasive approach for cancer detection and monitoring [121]. Among its diverse analyte repertoire, non-coding RNAs (ncRNAs)—including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs)—have garnered significant attention as potential biomarkers. These molecules, once considered transcriptional "noise," are now recognized as essential regulators of biological functions and exhibit high stability in circulation due to their protection within extracellular vesicles (e.g., exosomes) or protein complexes [122] [70] [123]. This Application Note details the validation protocols and analytical frameworks for implementing circulating ncRNAs as reliable biomarkers within the context of HCC research and clinical development.
Performance Metrics of Circulating ncRNA Biomarkers in HCC
Extensive research has quantified the diagnostic potential of various circulating ncRNAs, often demonstrating superior performance over traditional protein markers like AFP.
Table 1: Diagnostic Performance of Select Circulating miRNAs in HCC
| miRNA | Source | AUC | Sensitivity (%) | Specificity (%) | Reference |
|---|---|---|---|---|---|
| miR-21 | Plasma | 0.953 | 87.3 | 92.0 | [70] |
| miR-224 | Plasma | 0.940 | 92.5 | 90.0 | [70] |
| miR-122 | Plasma | 0.960 | 87.5 | 95.0 | [70] |
| miR-665 | Serum | 0.930 | 92.5 | 86.3 | [70] |
| miR-9-3p | Serum | - | 91.43 | 87.50 | [70] |
| miR-34a (Exosomal) | Serum | 0.664 | 78.3 | 51.7 | [70] |
| miR-34a + AFP | Serum | 0.855 | 68.3 | 93.3 | [70] |
The value of lncRNAs is also increasingly evident. For instance, the serum exosomal long ncRNA FOXD2-AS1 has demonstrated promising diagnostic potential in colorectal cancer, with an AUC of 0.758 for early-stage disease, highlighting the potential for similar applications in HCC [122]. Furthermore, exosomal lncRNA-GC1 distinguished gastric cancer patients from controls with AUCs exceeding 0.86, outperforming traditional markers like CEA and CA19-9 [122]. These findings underscore the broader potential of exosomal ncRNAs across cancer types.
Experimental Protocols for ncRNA Analysis from Liquid Biopsy
Robust and reproducible pre-analytical and analytical protocols are fundamental to successful biomarker validation. The following section outlines critical procedural steps.
Pre-Analytical Workflow: Sample Collection and Processing
The pre-analytical phase is critical for preserving ncRNA integrity and ensuring accurate downstream results.
Table 2: Comparison of Blood Collection Tubes for Liquid Biopsy
| Tube Type | Additive / Principle | Max Storage (RT) | Advantages / Considerations |
|---|---|---|---|
| K3EDTA | Chelating agent | ≤1 hour (4°C) | Standard, requires immediate processing [124]. |
| Streck Cell-Free DNA BCT | Chemical crosslinking | 14 days | Stabilizes nucleated cells, reduces gDNA contamination [124]. |
| PAXgene Blood ccfDNA Tube | Biological apoptosis prevention | 14 days | Inhibits leukocyte lysis and nuclease activity [124]. |
| Norgen cf-DNA/cf-RNA Preservative Tube | Osmotic cell stabilization | 30 days | Allows for concurrent isolation of cfDNA and cfRNA [124]. |
Recommended Protocol: Plasma Preparation
- Blood Collection: Draw blood into appropriate preservation tubes (e.g., Streck, PAXgene).
- Initial Centrifugation: Centrifuge within the manufacturer's stipulated time at 500-2,000 × g for 15-20 minutes at room temperature to separate cellular components from plasma [124].
- Secondary Centrifugation: Transfer the supernatant (plasma) to a new tube and perform a second, higher-speed centrifugation (e.g., 16,000 × g for 10 minutes) to remove residual cells and debris [124].
- Storage: Aliquot the clarified plasma to avoid freeze-thaw cycles and store at -80°C until nucleic acid extraction.
Analytical Workflow: ncRNA Isolation and Quantification
Protocol: Parallel Isolation of Cell-Free ncRNA and DNA For a multi-analyte approach, use a commercial kit designed for concurrent extraction of cfDNA and cfRNA (e.g., Norgen's kit) [124].
- Lysis: Add plasma to a lysis buffer to disrupt vesicles and release nucleic acids.
- Binding: Bind nucleic acids to a silica membrane column.
- Washing: Perform washes with appropriate buffers to remove contaminants.
- Elution: Elute cfDNA and cfRNA in separate fractions using nuclease-free water or elution buffer [124].
Protocol: ncRNA Quantification and Quality Control
- Quantification: Use fluorescence-based assays (e.g., Qubit RNA HS Assay) for accurate concentration measurement of low-abundance samples. Bioanalyzer or TapeStation systems provide integrity profiles [124].
- Reverse Transcription: Convert RNA to cDNA using reverse transcriptase with stem-loop primers for miRNAs or random hexamers/Oligo(dT) for lncRNAs and circRNAs.
- Quantification Methods:
- Quantitative RT-PCR (qRT-PCR): The gold standard for targeted, sensitive quantification of specific ncRNAs. Use TaqMan or SYBR Green chemistry [70].
- Droplet Digital PCR (ddPCR): Provides absolute quantification without a standard curve and is highly effective for detecting rare targets, offering superior precision for low-abundance ncRNAs [125] [124].
- Next-Generation Sequencing (NGS): Employ RNA-Seq for discovery-phase profiling, allowing for the identification of novel ncRNAs and differential expression analysis across the entire transcriptome [70] [123].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Kits for ncRNA Liquid Biopsy Workflows
| Item / Kit | Function | Key Characteristics |
|---|---|---|
| Streck Cell-Free DNA BCT | Blood collection & stabilization | Prevents leukocyte lysis and preserves cfNA profile for up to 14 days [124]. |
| Norgen cfNA Purification Kit | Parallel isolation of cfDNA & cfRNA | Enables multi-analyte analysis from a single plasma sample [124]. |
| Qiagen miRNeasy Serum/Plasma Kit | Selective isolation of small RNAs | Optimized for recovery of miRNA and other small RNAs. |
| TaqMan Advanced miRNA Assays | cDNA synthesis & qPCR of miRNAs | High sensitivity and specificity for mature miRNA targets. |
| Bio-Rad ddPCR Supermix | Absolute quantification of ncRNAs | Enables detection of rare targets without a standard curve [125]. |
| AGO2 Antibody | Immunoprecipitation of AGO2-bound ncRNAs | Isulates a specific population of circulating ncRNAs stabilized by Argonaute 2 protein [123]. |
| CD63/CD81 Antibodies | Immunocapture of exosomes | Enriches for exosomal populations carrying ncRNAs [122]. |
Regulatory Pathways and Functional Mechanisms of ncRNAs in HCC
Circulating ncRNAs often reflect the active regulatory processes within the tumor microenvironment. Understanding their functional mechanisms provides a biological rationale for their use as biomarkers.
Figure 1: Functional Mechanism of ncRNAs in HCC Progression. HCC cells release ncRNAs packaged in exosomes or other vesicles into circulation. Upon uptake by recipient cells (e.g., stromal, immune, or other tumor cells), these ncRNAs modulate key oncogenic signaling pathways, driving processes like metastasis and drug resistance [36] [126].
The diagram illustrates that validated circulating ncRNAs are not merely correlates of disease but are often functional mediators of HCC pathogenesis, underscoring their biological significance and reinforcing their value as biomarkers.
The integration of robust, standardized protocols for liquid biopsy handling with highly sensitive detection platforms like ddPCR and NGS positions circulating ncRNAs as formidable tools for the non-invasive monitoring of HCC. Their diagnostic performance, frequently surpassing traditional markers, and their direct involvement in tumorigenic pathways offer a dual rationale for their clinical translation. As research progresses, the validation of multi-ncRNA panels and their combination with other liquid biopsy analytes, such as ctDNA, will likely pave the way for their routine application in personalized oncology, ultimately improving early detection, treatment monitoring, and patient outcomes in HCC.
Conclusion
RNA sequencing has unequivocally established the critical role of ncRNAs in hepatocellular carcinoma, revealing a complex regulatory network that drives tumor initiation, progression, and therapy resistance. The integration of advanced methodologies, particularly single-cell sequencing and machine learning, is rapidly translating these discoveries into powerful diagnostic and prognostic tools that surpass traditional biomarkers. However, overcoming tumor heterogeneity and rigorously validating findings both computationally and experimentally remain vital challenges. Future research must focus on standardizing analytical pipelines, exploring the dynamic role of ncRNAs in the tumor immune microenvironment, and launching clinical trials for ncRNA-based therapeutics and liquid biopsies. Successfully bridging these gaps will pave the way for a new era of precision oncology in HCC management, ultimately improving patient outcomes.
References
- 1. MicroRNAs in the Pathogenesis of Hepatocellular Carcinoma [https://pmc.ncbi.nlm.nih.gov/articles/PMC7866004/]
- 2. Role of Non-Coding RNAs in Hepatocellular Carcinoma ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10647270/]
- 3. Function of the Long Noncoding RNAs in Hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9405066/]
- 4. ncRNA-mediated fatty acid metabolism reprogramming in ... [https://www.sciencedirect.com/science/article/abs/pii/S1043276023000383]
- 5. CircRNAs in hepatocellular carcinoma - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC9724243/]
- 6. Circular RNAs: characteristics, biogenesis, mechanisms and ... [https://jhoonline.biomedcentral.com/articles/10.1186/s13045-021-01145-8]
- 7. The role of long noncoding RNAs in hepatocellular carcinoma [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-020-01188-4]
- 8. Analysis of four long non-coding RNAs for hepatocellular ... [https://www.nature.com/articles/s41598-024-80926-w]
- 9. Establishing a miRNA panel for hepatocellular carcinoma ... [https://www.sciencedirect.com/science/article/pii/S2589004225012477]
- 10. Hepatocellular Carcinoma: The Role of MicroRNAs - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9138333/]
- 11. Integrative Analysis of Non-coding RNAs and Hippo ... [https://brieflands.com/journals/ijcm/articles/157228]
- 12. Increased MiR-221 expression in hepatocellular carcinoma ... [https://bmccancer.biomedcentral.com/articles/10.1186/1471-2407-13-21]
- 13. miR-221-3p regulates hepatocellular carcinoma cell ... [https://www.sciencedirect.com/science/article/pii/S1665268121002660]
- 14. Long non-coding RNA GAS5 in human cancer - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7400976/]
- 15. Oncogenic function of growth arrest-specific transcript 5 by ... [https://www.nature.com/articles/s12276-025-01459-4]
- 16. Long noncoding RNA GAS5 inhibits metastasis by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6356919/]
- 17. Long non-coding RNAs in hepatocellular carcinoma [https://pmc.ncbi.nlm.nih.gov/articles/PMC5583571/]
- 18. Non-coding RNA in cancer - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8564738/]
- 19. Targeting non-coding RNAs to overcome cancer therapy ... [https://www.nature.com/articles/s41392-022-00975-3]
- 20. A comprehensive insight into the correlation between ncRNAs ... [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-023-01092-6]
- 21. Emerging roles of non-coding RNAs in modulating the PI3K ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11406677/]
- 22. Non-coding RNAs and regulation of the PI3K signaling ... [https://www.sciencedirect.com/science/article/pii/S2468054024001574]
- 23. Cooperation between both Wnt/β-catenin and PTEN/PI3K/Akt ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3185965/]
- 24. RNA sequencing reveals the long noncoding RNA and ... [https://www.sciencedirect.com/science/article/pii/S0753332220303036]
- 25. The ncRNA-AURKA Interaction in Hepatocellular Carcinoma [https://pmc.ncbi.nlm.nih.gov/articles/PMC11595987/]
- 26. Hepatocellular carcinoma: signaling pathways and ... [https://www.nature.com/articles/s41392-024-02075-w]
- 27. Long non-coding RNA in hepatocellular carcinoma [https://pubmed.ncbi.nlm.nih.gov/41186893/]
- 28. Long non-coding RNA in hepatocellular carcinoma [https://link.springer.com/article/10.1007/s13402-025-01119-9]
- 29. Dynamic role of long noncoding RNA in liver diseases [https://pmc.ncbi.nlm.nih.gov/articles/PMC12078412/]
- 30. Non-coding RNAs as key regulators in hepatitis B virus- ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1602252/full]
- 31. Long non-coding RNA-based single and combination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12405140/]
- 32. Oncology Letters [https://www.spandidos-publications.com/10.3892/ol.2025.15356?text=fulltext]
- 33. Long non-coding RNAs and autophagy: dual drivers of ... [https://www.nature.com/articles/s41420-025-02667-7]
- 34. MiRNA delivery systems in cancer stem cell therapy [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04667-x]
- 35. Long non-coding rnas as key modulators of the immune ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1523190/full]
- 36. Non-coding RNAs in hepatocellular carcinoma: Insights ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2022.985815/full]
- 37. The Role of NcRNAs to Regulate Immune Checkpoints in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9019622/]
- 38. Non-coding RNA-based immunotherapy may improve the ... [https://www.sciencedirect.com/science/article/pii/S0753332223008958]
- 39. Experimental Models of Hepatocellular Carcinoma—A ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8344976/]
- 40. The role of long non-coding RNA in hepatocellular carcinoma [https://pmc.ncbi.nlm.nih.gov/articles/PMC10929815/]
- 41. protocol for a prospective cohort study of high-risk patients for ... [https://bmccancer.biomedcentral.com/articles/10.1186/s12885-023-11167-9]
- 42. Dual tissue mRNA and serum protein signatures improve ... [https://www.nature.com/articles/s41698-025-01142-x]
- 43. Estimation of sample size and outcome evaluation [https://bio-protocol.org/exchange/minidetail?id=9665959&type=30]
- 44. Single-cell RNA sequencing analysis reveals potential key ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11618547/]
- 45. Sample size calculation [https://www.sciencedirect.com/science/article/abs/pii/S0301054613001523]
- 46. Integrating single-cell RNA-Seq and bulk RNA-Seq data to ... [https://www.nature.com/articles/s41598-025-85506-0]
- 47. An Update of Immunohistochemistry in Hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12428263/]
- 48. A single-cell atlas of the multicellular ecosystem of primary ... [https://www.nature.com/articles/s41467-022-32283-3]
- 49. Special Issue “Regulation by Non-Coding RNAs 2025†[https://www.mdpi.com/1422-0067/26/22/10885]
- 50. A comprehensive genome-wide analysis of long ... [https://pubmed.ncbi.nlm.nih.gov/29047230/]
- 51. From bulk, single-cell to spatial RNA sequencing [https://www.nature.com/articles/s41368-021-00146-0]
- 52. Understanding Bulk and Single-Cell RNA Sequencing [https://www.cd-genomics.com/resource-bulk-rna-sequencing-vs-single-cell-rna-sequencing.html]
- 53. Bulk RNA Sequencing versus Single-Cell RNAseq: 2 Powerful... [https://sampled.com/bulk-rna-sequencing-vs-single-cell-rna-sequencing/]
- 54. Profiling, clinicopathological correlation and functional ... [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-017-0733-5]
- 55. Single cell RNA-seq analysis identifies a noncoding RNA ... [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-021-01473-w]
- 56. Integrative Analysis of scRNA-Seq and Bulk RNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11878290/]
- 57. Integration of single-cell and bulk RNA sequencing data to ... [https://www.nature.com/articles/s41598-025-20691-6]
- 58. A Guide to Basic RNA Sequencing Data Processing and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12067304/]
- 59. [Tutorial] Bulk RNA-seq DE analysis [https://informatics.fas.harvard.edu/resources/tutorials/differential-expression-analysis/]
- 60. A Beginner's Guide to Analysis of RNA Sequencing Data [https://pmc.ncbi.nlm.nih.gov/articles/PMC6096346/]
- 61. Hepatocellular Carcinoma Pipeline Drugs Report 2025 [https://www.barchart.com/story/news/34928227/hepatocellular-carcinoma-pipeline-drugs-report-2025-exploring-novel-mechanisms-of-action-ongoing-clinical-trials-pharma-collaborations-and-the-future-outlook]
- 62. Integration of single-cell and bulk RNA sequencing identifies ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0322706]
- 63. Prognostic model for predicting recurrence‑free survival in ... [https://www.spandidos-publications.com/10.3892/ol.2025.15283]
- 64. Integrative Single-Cell and Bulk Transcriptomic Analysis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12562699/]
- 65. Integrated analysis of single-cell and bulk RNA sequencing ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2024.1407428/full]
- 66. Integrating single-cell RNA sequencing and artificial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12405525/]
- 67. Multi-omics and single-cell approaches reveal molecular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12138379/]
- 68. Current Status and Analysis of Machine Learning in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10412715/]
- 69. Decoding the molecular and genomic landscape of ... [https://www.nature.com/articles/s44355-025-00038-8]
- 70. Circulating Non-Coding RNAs as Potential Diagnostic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9484560/]
- 71. Identifying novel transcript biomarkers for hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8394105/]
- 72. Minimal clinical predictors enable machine learning ... [https://www.nature.com/articles/s41598-025-22927-x]
- 73. Machine learning-based identification of diagnostic and ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0331118]
- 74. Machine learning-based identification of diagnostic and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12393733/]
- 75. Multiple machine learning algorithms identified SLC6A8 as ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12003237/]
- 76. Integrating single-cell RNA sequencing and artificial ... [https://www.nature.com/articles/s41698-025-00952-3]
- 77. Highly effective batch effect correction method for RNA-seq ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11718288/]
- 78. Batch Correction Analysis | Griffith Lab [https://rnabio.org/module-03-expression/0003/06/02/Batch-Correction/]
- 79. Batch effect detection and correction in RNA-seq data using ... [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-022-04775-y]
- 80. a practical guide to RNA sequencing data analysis [https://pmc.ncbi.nlm.nih.gov/articles/PMC12599993/]
- 81. A comprehensive workflow for optimizing RNA-seq data analysis [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-024-10414-y]
- 82. Identification of novel long non-coding RNAs deregulated in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5077982/]
- 83. Exploration of heterogeneity and recurrence signatures in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12330942/]
- 84. Machine learning for catalysing the integration of noncoding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11314885/]
- 85. A Review of Feature Selection Methods for Machine Learning ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9580915/]
- 86. Single‑cell RNA sequencing data dimensionality reduction ... [https://www.spandidos-publications.com/10.3892/wasj.2025.315]
- 87. Optimal features selection in the high dimensional data ... [https://www.sciencedirect.com/science/article/pii/S2405844024132720]
- 88. Optimizing high dimensional data classification with a ... [https://www.nature.com/articles/s41598-025-08699-4]
- 89. Integrating bulk and single-cell RNA sequencing data to ... [https://www.sciencedirect.com/science/article/pii/S2405844023092915]
- 90. Machine-Learning-Based Identification of Key Feature ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11268357/]
- 91. ncFN: a comprehensive non-coding RNA function ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12550578/]
- 92. Pathway enrichment analysis and visualization of omics data ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6607905/]
- 93. Chapter 15 Visualization of functional enrichment result [https://yulab-smu.top/biomedical-knowledge-mining-book/enrichplot.html]
- 94. Comprehensive analysis of LncRNA-miRNA-mRNA ... [https://www.nature.com/articles/s41598-025-20187-3]
- 95. Harnessing non-coding RNA therapeutics for ... [https://pubmed.ncbi.nlm.nih.gov/41238082/]
- 96. Public RNA-Seq and scRNA-Seq Databases [https://bigomics.ch/blog/ultimate-guide-to-public-rnaseq-and-sc-rna-seq-databases/]
- 97. Identification and validation of feature genes in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12558189/]
- 98. RNA sequencing analysis of hepatocellular carcinoma ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9315135/]
- 99. from discovery and functional prediction to clinical application [https://jhoonline.biomedcentral.com/articles/10.1186/s13045-020-00945-8]
- 100. Integrative approaches for studying the role of noncoding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9117541/]
- 101. Firalink: A bioinformatics pipeline for long non-coding RNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10523182/]
- 102. Integrative in Silico and in vitro validation suggest ... [https://www.nature.com/articles/s41598-025-08777-7]
- 103. Functional Screening Techniques to Identify Long Non ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7763270/]
- 104. In vivo functional analysis of non-conserved human ... [https://www.nature.com/articles/s41467-019-13688-z]
- 105. Prognostic model for predicting recurrence‑free survival in... [https://www.spandidos-publications.com/10.3892/ol.2025.15283?text=fulltext]
- 106. Single-Cell RNA Sequencing Integrated with Bulk-RNA ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12318859/]
- 107. Frontiers | Establishment of a nomogram-based prognostic model... [https://www.frontiersin.org/journals/molecular-biosciences/articles/10.3389/fmolb.2025.1561114/full]
- 108. Construction of an interpretable model for predicting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11450703/]
- 109. Risk scoring based on expression of long non-coding ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7108035/]
- 110. Development and validation of an Immune-related Gene ... [https://www.nature.com/articles/s41598-025-90183-0]
- 111. Cuproptosis-related long non - coding model that effectively... RNAs [https://pubmed.ncbi.nlm.nih.gov/36310708/]
- 112. Transcriptome sequencing of hepatocellular carcinoma ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9484539/]
- 113. A comparison of clinical pathologic characteristics between ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9034573/]
- 114. A novel disulfidptosis-related lncRNA signature for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12582836/]
- 115. The outcomes and prognostic factors of patients with ... [https://www.sciencedirect.com/science/article/pii/S0929664622004284]
- 116. RNA sequencing data of HCC tissues [https://bio-protocol.org/exchange/minidetail?id=3287055&type=30]
- 117. Machine learning-based hepatocellular carcinoma risk ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12504168/]
- 118. Non-coding RNAs in Hepatocellular Carcinoma [https://www.genesispub.org/non-coding-rnas-in-hepatocellular-carcinoma]
- 119. Radiomics-Driven Machine Learning Models for Non ... [https://www.sciencedirect.com/science/article/abs/pii/S0009926025003654]
- 120. HTRecNet: a deep learning study for efficient and accurate ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1549811/full]
- 121. Liquid biopsy in cancer: current status, challenges and ... [https://www.nature.com/articles/s41392-024-02021-w]
- 122. Exosomal ncRNAs in liquid biopsy: a new paradigm for ... [https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2025.1615433/full]
- 123. Proceedings of cell-free noncoding RNA biomarker studies in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12495990/]
- 124. From Sampling to Sequencing: A Liquid Biopsy Pre-Analytic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8232701/]
- 125. Drug Discovery News — ctDNA monitoring is providing a ... [https://friendsofcancerresearch.org/news/drug-discovery-news-ctdna-monitoring-is-providing-a-smarter-way-to-track-and-treat-cancer/]
- 126. Non-coding RNAs in hepatocellular carcinoma metastasis [https://www.sciencedirect.com/science/article/abs/pii/S0010482524009521]