Revolutionizing HCC Diagnosis: Machine Learning Integration of lncRNA Biomarkers
Hepatocellular carcinoma (HCC) remains a leading cause of cancer mortality globally, largely due to limitations in early detection.
Revolutionizing HCC Diagnosis: Machine Learning Integration of lncRNA Biomarkers
Abstract
Hepatocellular carcinoma (HCC) remains a leading cause of cancer mortality globally, largely due to limitations in early detection. This article explores the transformative integration of machine learning (ML) with long non-coding RNA (lncRNA) biomarkers to address this critical diagnostic challenge. We provide a comprehensive analysis for researchers and drug development professionals, covering the foundational biology of lncRNAs in HCC, advanced methodological approaches for ML model development, strategies for troubleshooting and optimizing diagnostic signatures, and rigorous validation frameworks. The synthesis of current evidence demonstrates that ML-driven lncRNA panels significantly outperform traditional biomarkers like AFP, achieving diagnostic accuracies exceeding 98% in recent studies. This paradigm shift promises to enable non-invasive, cost-effective, and highly precise tools for early HCC detection, prognosis prediction, and personalized therapeutic guidance, ultimately paving the way for improved patient outcomes in precision oncology.
The Biology of lncRNAs in Hepatocellular Carcinoma: From Molecular Mechanisms to Diagnostic Potential
Definition and Fundamental Characteristics of Long Non-Coding RNAs
Long non-coding RNAs (lncRNAs) are broadly defined as RNA transcripts exceeding 200 nucleotides in length that lack protein-coding potential [1] [2]. This operational definition originated from biochemical purification protocols that separate these longer RNAs from infrastructural RNAs like tRNAs, snRNAs, and snoRNAs [1]. The human genome encodes a vast repertoire of lncRNAs, with current annotations estimating between 20,000 to over 90,000 lncRNA genes, potentially outnumbering protein-coding genes [3] [2].
LncRNAs exhibit several distinctive features compared to messenger RNAs (mRNAs). While many are RNA polymerase II (Pol II) transcribed, 5'-capped, and polyadenylated, a significant subset lacks poly(A) tails [1] [2]. They generally display lower sequence conservation, contain fewer and longer exons, and undergo less efficient splicing with more non-canonical splice sites [3] [4]. LncRNAs are typically expressed at lower levels than protein-coding genes and show remarkably precise tissue-specific, cell-type-specific, and developmental-stage-specific expression patterns, making them particularly attractive for diagnostic applications [3] [4].
Table 1: Key Characteristics of Long Non-Coding RNAs
| Feature | Description | Biological Significance |
|---|---|---|
| Length | >200 nucleotides | Distinguishes from small non-coding RNAs (miRNAs, siRNAs) [1] |
| Coding Potential | Non-protein-coding | Primary function is regulatory rather than template for translation [3] |
| Expression Level | Generally low abundance | Requires sensitive detection methods; reduces transcriptional burden [4] [5] |
| Expression Pattern | Highly cell-type and developmental stage-specific | Ideal for tissue-specific regulation and as disease-specific biomarkers [3] [6] |
| Sequence Conservation | Lower than protein-coding genes | Function may be conserved through structures/motifs rather than primary sequence [3] [4] |
| Subcellular Localization | Often nuclear enriched | Reflects roles in chromatin regulation and transcription [4] |
Diverse Functional Roles in Gene Regulation
LncRNAs function as versatile regulators of gene expression through mechanisms correlated with their subcellular localization. Their functional diversity stems from ability to interact with DNA, RNA, and proteins through specific structural domains [4] [7].
Nuclear Functions
In the nucleus, lncRNAs orchestrate epigenetic regulation by recruiting chromatin-modifying complexes to specific genomic loci. For example, XIST initiates X-chromosome inactivation by coating the future inactive X chromosome and recruiting repressive complexes, while HOTAIR recruits Polycomb Repressive Complex 2 (PRC2) to silence tumor suppressor genes, promoting cancer metastasis [3] [4]. LncRNAs also regulate transcription by influencing transcription factor activity or RNA polymerase II recruitment, and some act as enhancer RNAs (eRNAs) to stimulate transcription of nearby genes [4] [7].
Cytoplasmic Functions
In the cytoplasm, lncRNAs influence mRNA stability, translation, and post-translational modifications. They can act as competing endogenous RNAs (ceRNAs) that "sponge" miRNAs, preventing them from repressing their target mRNAs [3]. Some lncRNAs directly interact with mRNA transcripts or proteins to modulate their stability and translation, while others participate in cellular signaling pathways [4] [7].
Experimental Protocols for lncRNA Investigation in Cancer Research
Protocol: Identification of Diagnostic lncRNA Biomarkers Using Machine Learning
This protocol outlines the workflow for discovering lncRNA biomarkers for hepatocellular carcinoma (HCC) diagnosis by integrating high-throughput transcriptomic data with machine learning approaches [8] [6].
Step 1: Sample Collection and RNA Sequencing
- Collect matched tumor and normal tissue samples from HCC patients and controls. Plasma or serum can be used for liquid biopsy approaches [6].
- Extract total RNA using kits designed to preserve long RNA species (e.g., miRNeasy Mini Kit).
- Perform stranded total RNA sequencing with rRNA depletion (not poly-A selection) to capture both polyadenylated and non-polyadenylated lncRNAs. Use UMI barcodes to eliminate PCR duplicates [2].
Step 2: Bioinformatics Processing
- Align sequencing reads to the reference genome using splice-aware aligners (STAR, HISAT2).
- Quantify lncRNA expression using comprehensive annotations (GENCODE, NONCODE).
- Identify differentially expressed lncRNAs (adjusted p-value < 0.05, |logFC| > 1) between tumor and normal groups [8].
Step 3: Machine Learning Feature Selection
- Apply Support Vector Machine-Recursive Feature Elimination (SVM-RFE) and Random Forest-Recursive Feature Elimination (RF-RFE) to identify the most informative diagnostic lncRNAs.
- Perform 10-fold cross-validation with multiple iterations (≥50) to ensure robust feature selection.
- Select lncRNAs consistently chosen in >90% of iterations for final model training [8].
Step 4: Model Validation
- Validate model performance on independent datasets using AUC (Area Under the Curve), sensitivity, specificity, and accuracy metrics.
- Perform permutation testing (n=100) to confirm that observed performance exceeds null distributions [8] [6].
Protocol: Functional Validation of Candidate lncRNAs in HCC
Step 1: Knockdown Using Lincode siRNAs
- Design siRNA pools targeting candidate lncRNAs (e.g., LINC00152, UCA1, HOTAIR).
- Transfert HCC cell lines (HepG2, Huh7) using appropriate transfection reagents.
- Confirm knockdown efficiency (>70%) after 48-72 hours using qRT-PCR with lncRNA-specific primers [5].
Step 2: Phenotypic Assays
- Assess proliferation changes using MTT or CellTiter-Glo assays.
- Evaluate apoptosis by flow cytometry with Annexin V/PI staining.
- Measure invasion capacity through Transwell Matrigel invasion assays [3] [6].
Step 3: Mechanistic Studies
- Determine subcellular localization by RNA fluorescence in situ hybridization (RNA-FISH).
- Identify interacting partners by RNA immunoprecipitation (RIP) or CLIP-seq.
- Investigate effects on candidate target genes by qRT-PCR and western blot [3] [5].
Table 2: Key Research Reagent Solutions for lncRNA Functional Studies
| Reagent Type | Specific Product Examples | Application in lncRNA Research |
|---|---|---|
| siRNA for Knockdown | Lincode siRNA pools [5] | Effective lncRNA knockdown with predesigned human and mouse reagents |
| CRISPR Tools | CRISPR-Cas9 guide RNAs [5] | lncRNA gene knockout or modification through genomic editing |
| qRT-PCR Kits | PowerTrack SYBR Green Master Mix [6] | Sensitive quantification of lncRNA expression levels |
| RNA Extraction Kits | miRNeasy Mini Kit [6] | Preserves long RNA species while also capturing small RNAs |
| Sequencing Kits | NEXTFLEX Rapid Directional RNA-Seq [2] | Strand-specific library prep for accurate lncRNA transcript quantification |
| Lentiviral Systems | shMIMIC Inducible Lentiviral microRNA [5] | Inducible expression systems for difficult-to-transfect cells |
lncRNAs as Biomarkers in Hepatocellular Carcinoma
LncRNAs show exceptional promise as diagnostic and prognostic biomarkers in HCC due to their tissue-specific expression, deregulation in cancer, and detectability in liquid biopsies [6]. Several lncRNAs have been identified as particularly relevant to HCC pathogenesis and clinical management.
Table 3: Diagnostic Performance of Selected lncRNAs in Hepatocellular Carcinoma
| lncRNA | Expression in HCC | Biological Function in HCC | Diagnostic Performance |
|---|---|---|---|
| LINC00152 | Upregulated | Promotes cell proliferation through regulation of CCDN1 [6] | AUC: 0.83, Sensitivity: 83%, Specificity: 67% [6] |
| UCA1 | Upregulated | Enhances proliferation and inhibits apoptosis [6] | AUC: 0.77, Sensitivity: 60%, Specificity: 53% [6] |
| GAS5 | Downregulated | Tumor suppressor; activates CHOP and caspase-9 pathways [6] | - |
| LINC00853 | Upregulated | Potential oncogenic functions [6] | - |
| HOTAIR | Upregulated | Promotes metastasis; independent predictor of poor survival [3] | Associated with poor overall and disease-free survival [3] |
| Machine Learning Panel | Combined signature | Integration of multiple lncRNAs with conventional biomarkers [6] | Sensitivity: 100%, Specificity: 97% [6] |
The combination of multiple lncRNAs into diagnostic panels significantly enhances performance compared to individual markers. When LINC00152, LINC00853, UCA1, and GAS5 were integrated with conventional laboratory parameters (AFP, ALT, AST) using machine learning algorithms, the model achieved 100% sensitivity and 97% specificity for HCC detection, substantially outperforming individual lncRNAs or AFP alone [6]. The LINC00152 to GAS5 expression ratio has emerged as a particularly promising prognostic indicator, with higher ratios correlating with increased mortality risk [6].
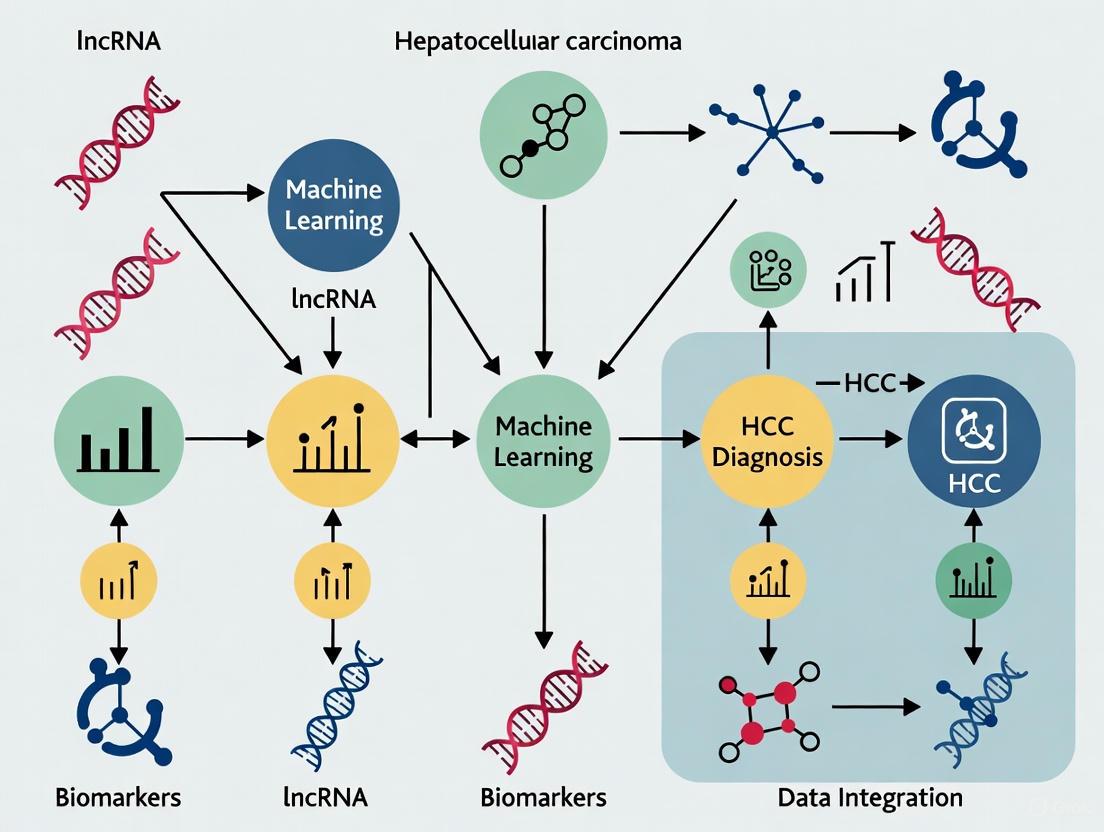
Integration of lncRNAs into Machine Learning Frameworks for HCC Diagnosis
Machine learning approaches are revolutionizing lncRNA biomarker development by enabling analysis of complex expression patterns that elude conventional statistical methods [9] [8]. The integration of lncRNA data into ML pipelines follows a structured approach:
Feature Selection Methods
- SVM-RFE (Support Vector Machine-Recursive Feature Elimination): Effectively identifies biologically relevant lncRNA features by iteratively removing the least important features [8].
- RF-RFE (Random Forest-Recursive Feature Elimination): Combines ensemble learning with recursive feature elimination for robust feature selection [8].
- LASSO (Least Absolute Shrinkage and Selection Operator): Performs variable selection and regularization to enhance prediction accuracy and interpretability, particularly for prognostic models [8].
Model Performance and Validation In HCC diagnostics, ML models trained on lncRNA expression data have demonstrated exceptional performance. One study achieved AUC = 1.0 in the training set (TCGA), with strong generalizability to external validation sets (AUC = 0.95 and 0.879) [8]. Permutation testing confirmed these results were statistically significant beyond null distributions [8].
Multi-Omics Integration The most powerful predictive models integrate lncRNA data with other molecular features and clinical parameters. This includes combining lncRNA expression with:
- mRNA expression profiles of key cell cycle regulators [8]
- Conventional serum biomarkers (AFP, ALT, AST) [6]
- Clinical staging and histopathological grading [8] [6]
This integrated approach facilitates the development of comprehensive diagnostic and prognostic signatures that more accurately reflect the molecular complexity of hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) represents a major global health challenge, ranking as the sixth most diagnosed cancer and the third leading cause of cancer-related deaths worldwide [10]. The pathogenesis of HCC involves complex biological processes including DNA damage, epigenetic modifications, and oncogene mutations, with long non-coding RNAs (lncRNAs) emerging as crucial regulators [11]. These RNA molecules, exceeding 200 nucleotides in length without protein-coding capacity, are intensively involved in HCC occurrence, metastasis, and progression through diverse mechanisms including miRNA sponging, chromatin remodeling, and protein interactions [12] [11].
LncRNAs demonstrate remarkable tissue and cellular specificity, making them ideal candidates for biomarker development. Their expression is regulated by various epigenetic mechanisms including DNA methylation, histone modifications, and RNA modifications, creating a complex regulatory network that influences HCC pathogenesis [10]. The dual role of lncRNAs as both oncogenic drivers and tumor suppressors presents a promising frontier for precision diagnostics and innovative therapeutics in HCC management, particularly when integrated with machine learning approaches for biomarker discovery and validation.
Molecular Mechanisms of Dysregulated lncRNAs in HCC
Oncogenic lncRNAs and Their Pathways
Oncogenic lncRNAs promote HCC development and progression through various mechanisms. They inhibit apoptosis, enhance cell survival by interacting with chromatin modifiers, alter DNA methylation or histone modifications, and promote oncogene expression while repressing tumor suppressor genes [13]. For instance, silencing lncRNA SLC7A11-AS1 effectively suppresses HCC progression, as confirmed by both in vivo and in vitro experiments [13]. METTL3 facilitates m6A modification of SLC7A11-AS1, enhancing its expression in HCC. Subsequently, SLC7A11-AS1 downregulates KLF9 by influencing STUB1-mediated ubiquitination degradation, allowing KLF9 to elevate PHLPP2 expression, resulting in AKT pathway inactivation [13].
The lncRNA HOMER3-AS1 shows elevated levels in HCC and is associated with increased tumor growth, migration, invasion, and poor patient survival. It contributes to recruitment and polarization of M2 macrophages, further facilitating cancer cell proliferation [13]. Another significant oncogenic lncRNA, SNHG6, operates as a competitive endogenous RNA (ceRNA), binding to miR-204-5p to increase E2F1 expression and promote the G1-S phase transition, driving HCC tumorigenesis [13].
Table 1: Key Oncogenic lncRNAs in HCC and Their Mechanisms
| LncRNA | Expression in HCC | Molecular Mechanism | Functional Outcome |
|---|---|---|---|
| SLC7A11-AS1 | Upregulated | METTL3-mediated m6A modification; downregulates KLF9 | AKT pathway inactivation; promotes progression |
| HOMER3-AS1 | Upregulated | Recruitment and polarization of M2 macrophages | Enhanced growth, migration, invasion |
| SNHG6 | Upregulated | Sponges miR-204-5p to increase E2F1 | G1-S phase transition; tumorigenesis |
| CCAT2 | Upregulated | Inhibits miR-145 maturation; regulates miR-4496/Atg5 axis | Proliferation and metastasis |
| HOTAIR | Upregulated | Decreases miR-122 via DNMTs-induced DNA methylation | Cyclin G1 dysregulation; sorafenib resistance |
| H19 | Upregulated | Downregulates miRNA-15b, activates CDC42/PAK1 axis | Increased proliferation rate |
| HULC | Upregulated | Multiple mechanisms in different contexts | Proliferation, migration, apoptosis regulation |
| NEAT1 | Upregulated | Various oncogenic pathways | Proliferation, migration, apoptosis regulation |
Tumor Suppressor lncRNAs and Their Functions
Tumor suppressor lncRNAs play protective roles against HCC development and progression. The lncRNA GAS5 (growth arrest-specific 5) acts as a tumor suppressor by triggering CHOP and caspase-9 signal pathways, thereby inhibiting cancer cell proliferation and activating apoptosis [6]. Another significant tumor suppressor, MEG3 (maternally expressed 3), demonstrates reduced expression in HCC due to promoter region hypermethylation [10]. Treatment of HCC cell lines with decitabine or silencing of DNMT1/3b leads to substantial up-regulation of MEG3 expression, which enhances apoptosis and impedes HCC cell proliferation [10].
The regulatory dynamics of tumor suppressor lncRNAs often involve polymorphic variations. For instance, a 5-base pair indel polymorphism (rs145204276) in the GAS5 promoter region shows a strong association between the deletion allele and increased GAS5 expression, as well as heightened methylation of a neighboring CpG site within the promoter region [10]. This highlights the complex epigenetic regulation governing tumor suppressor lncRNA expression in HCC.
Table 2: Key Tumor Suppressor lncRNAs in HCC and Their Mechanisms
| LncRNA | Expression in HCC | Molecular Mechanism | Functional Outcome |
|---|---|---|---|
| GAS5 | Downregulated | Triggers CHOP and caspase-9 signal pathways | Inhibits proliferation, activates apoptosis |
| MEG3 | Downregulated | Promoter hypermethylation; regulated by DNMT1/3b | Enhances apoptosis, impedes proliferation |
| LINC00153 | Context-dependent | Part of diagnostic panels with UCA1 and AFP | Potential tumor suppressor in specific contexts |
| LINC00853 | Context-dependent | Used in machine learning diagnostic models | Potential tumor suppressor in specific contexts |
LncRNAs in Autophagy and ER Stress Regulation
The interplay between lncRNAs and cellular stress responses represents a critical aspect of HCC pathogenesis. Autophagy, a conserved catabolic pathway essential for cellular homeostasis, plays a paradoxical role in HCC—acting as a tumor suppressor during initiation but promoting survival and progression in advanced stages [12]. Long non-coding RNAs have emerged as critical regulators of autophagy, influencing tumorigenesis, metastasis, and therapy resistance through integration into key signaling networks such as PI3K/AKT/mTOR, AMPK, and Beclin-1 [12].
Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) also interact significantly with lncRNAs in HCC. Under stressful conditions, tumor cells activate adaptive mechanisms like ER stress due to increased demand for protein biosynthesis [13]. The intensity and duration of UPR dictates the cells' pro-survival and pro-apoptotic fate, with lncRNAs serving as key epigenetic modifiers in this process [13]. Dysregulated lncRNAs contribute to various facets of HCC, including apoptosis resistance, enhanced proliferation, invasion, and metastasis, all driven by ER stress responses.
Machine Learning Approaches for lncRNA Biomarker Integration
Diagnostic Model Development
Machine learning algorithms have demonstrated remarkable efficacy in integrating lncRNA biomarkers for HCC diagnosis. One study developed a model incorporating four lncRNAs (LINC00152, LINC00853, UCA1, and GAS5) with conventional laboratory parameters, achieving 100% sensitivity and 97% specificity in HCC diagnosis [6]. While individual lncRNAs showed moderate diagnostic accuracy with sensitivity and specificity ranging from 60-83% and 53-67% respectively, the integrated machine learning approach significantly outperformed single-marker analyses [6].
Another research effort employed five classifiers (KNN, RF, SVM, LGBM, and DNNs) to predict HCC using a 22-feature set that included RQLnc-WRAP53 and RQLncRNA-RP11-513I15.6 [14]. The Light Gradient Boosting Machine (LGBM) achieved the highest accuracy of 98.75% in predicting HCC, surpassing Random Forest (96.25%), DNN (91.25%), SVC (88.75%), and KNN (87.50%) [14]. This demonstrates the power of ensemble methods in handling complex lncRNA expression patterns for diagnostic applications.
Prognostic Signature Development
Machine learning has also enabled the development of robust prognostic signatures for HCC recurrence prediction. One study constructed a 4-lncRNA signature consisting of AC108463.1, AF131217.1, CMB9-22P13.1, and TMCC1-AS1 for predicting HCC early recurrence [15]. The construction process involved three machine learning methods—LASSO, Random Forest, and SVM-Recursive Feature Elimination—to identify the most predictive lncRNA combinations from initial candidate pools [15].
When combined with AFP and TNM staging systems, this 4-lncRNA signature demonstrated excellent predictability for HCC early recurrence. Patients in the high-risk group showed significantly higher early recurrence rates compared to those in the low-risk group [15]. Furthermore, antitumor immune cells, including activated B cells, type 1 T helper cells, natural killer cells, and effective memory CD8 T cells, were enriched in patients with low-risk HCCs, providing mechanistic insights into the differential recurrence rates [15].
Table 3: Machine Learning-Derived lncRNA Signatures in HCC
| Study | lncRNA Signature | ML Algorithms Used | Performance | Application |
|---|---|---|---|---|
| Elsayed et al. [6] | LINC00152, LINC00853, UCA1, GAS5 | Python's Scikit-learn platform | 100% sensitivity, 97% specificity | HCC diagnosis |
| Noureldeen et al. [14] | RQLnc-WRAP53, RQLncRNA-RP11-513I15.6 | LGBM, RF, DNN, SVC, KNN | 98.75% accuracy (LGBM) | HCC diagnosis |
| Zhou et al. [15] | AC108463.1, AF131217.1, CMB9-22P13.1, TMCC1-AS1 | LASSO, RF, SVM-RFE | Excellent early recurrence prediction | Prognostic stratification |
Experimental Protocols for lncRNA Analysis
Sample Collection and RNA Isolation
Protocol: Plasma Sample Collection and RNA Extraction
Sample Collection: Collect plasma samples from HCC patients and age-matched healthy controls. For HCC patients, samples can be retrieved from hospital biobanks, while control samples should be collected following standard protocols [6]. All participants must provide written informed consent, and the study protocol should be approved by the institutional ethical committee.
RNA Isolation: Isolate total RNA using the miRNeasy Mini Kit (QIAGEN, cat no. 217004) according to the manufacturer's protocol [6]. This kit efficiently recovers both long and short RNA species, ensuring comprehensive lncRNA analysis.
Quality Control: Validate RNA quality and purity using a Qubit 3.0 Fluorimeter with appropriate RNA assay kits [14]. Ensure RNA integrity numbers (RIN) exceed 7.0 for reliable downstream applications.
cDNA Synthesis: Perform reverse transcription into complementary DNA using the RevertAid First Strand cDNA Synthesis Kit [6]. Use a thermal cycler programmed according to the manufacturer's specifications, typically involving incubation at 42°C for 60 minutes followed by enzyme inactivation at 70°C for 5 minutes.
Quantitative Real-Time PCR Analysis
Protocol: qRT-PCR for lncRNA Quantification
Primer Design: Utilize commercially available primer sequences designed by established companies such as Thermo Fisher Scientific [6]. Validate primer specificity through melt curve analysis and gel electrophoresis.
Reaction Setup: Employ PowerTrack SYBR Green Master Mix kit and a ViiA 7 real-time PCR system for quantification [6]. Set up reactions in triplicate to ensure technical reproducibility.
Thermal Cycling Conditions: Program the qRT-PCR instrument with the following standard conditions: initial denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds, and annealing/extension at 60°C for 1 minute [6].
Data Normalization: Use housekeeping genes such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or GAD1 for normalization of expression data [6] [14]. Calculate relative expression using the ΔΔCT method, with results expressed as fold changes relative to control samples.
Machine Learning Implementation
Protocol: Development of lncRNA-Based Diagnostic Models
Feature Selection: Identify differentially expressed lncRNAs through RNA sequencing analysis of HCC and adjacent normal tissues [15]. Apply multiple differential expression analysis methods (DESeq2, edgeR, limma) with cutoff values of |log2FC| > 1 and FDR < 0.05 [15].
Data Preprocessing: Normalize expression data, handle missing values, and partition datasets into training and validation cohorts (typically 70:30 ratio) [15]. Ensure representative sampling across clinical stages and etiologies.
Model Training: Implement multiple machine learning algorithms including Random Forest, Support Vector Machines, Light Gradient Boosting Machines, and Deep Neural Networks [14]. Use k-fold cross-validation (typically 5-10 folds) to optimize hyperparameters and prevent overfitting.
Model Validation: Evaluate model performance on independent validation cohorts using metrics including accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve [6] [14]. Compare model performance against established clinical biomarkers like AFP.
Visualization of Key Pathways and Workflows
LncRNA Biogenesis and Functional Mechanisms
LncRNA Biogenesis and Functional Mechanisms in HCC
Machine Learning Workflow for lncRNA Biomarker Development
ML Workflow for lncRNA Biomarker Development
Research Reagent Solutions
Table 4: Essential Research Reagents for lncRNA Studies in HCC
| Reagent Category | Specific Product/Kit | Manufacturer | Application Purpose | Key Features |
|---|---|---|---|---|
| RNA Extraction | miRNeasy Mini Kit | QIAGEN (cat no. 217004) | Total RNA isolation from plasma/serum | Efficient recovery of long and short RNAs |
| cDNA Synthesis | RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific (cat no. K1622) | Reverse transcription for qRT-PCR | High efficiency for lncRNA templates |
| qRT-PCR Master Mix | PowerTrack SYBR Green Master Mix | Applied Biosystems (cat no. A46012) | lncRNA quantification | Sensitive detection with low background |
| qRT-PCR System | ViiA 7 Real-Time PCR System | Applied Biosystems | High-throughput lncRNA expression | Multi-well format for screening panels |
| RNA Quality Control | Qubit RNA HS Assay Kit | Invitrogen (Cat. no. Q32852) | RNA quantification and quality assessment | Accurate concentration measurements |
| PCR Primers | Custom LNA Primer Assays | Various suppliers | Specific lncRNA detection | Enhanced specificity for lncRNA targets |
| Methylation Analysis | EZ DNA Methylation Kit | Zymo Research | Promoter methylation studies | Bisulfite conversion for epigenetic analysis |
| Machine Learning | Scikit-learn Platform | Python Open Source | Diagnostic model development | Comprehensive ML algorithm library |
The integration of lncRNA biology with machine learning approaches represents a paradigm shift in HCC research and clinical practice. Dysregulated lncRNAs serve as critical drivers of hepatocarcinogenesis through diverse mechanisms, while their tissue specificity and detectability in liquid biopsies make them ideal biomarker candidates. The remarkable performance of machine learning models incorporating lncRNA signatures—achieving up to 98.75% accuracy in HCC diagnosis—underscores the transformative potential of this integrated approach [14].
Future directions should focus on validating these findings in larger, multi-center cohorts and addressing technical challenges related to sample processing, standardization, and analytical variability. Furthermore, the therapeutic targeting of oncogenic lncRNAs using approaches such as antisense oligonucleotides, siRNAs, or CRISPR/Cas systems presents an exciting frontier for HCC treatment [12]. As our understanding of lncRNA biology deepens and machine learning algorithms become more sophisticated, the integration of these fields promises to revolutionize HCC management through improved early detection, accurate prognosis prediction, and personalized therapeutic interventions.
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor worldwide and represents the third leading cause of cancer-related deaths, with a dismal 5-year survival rate of approximately 5%-6% [16] [17]. The molecular pathogenesis of HCC is complex, and recent research has shifted focus toward non-coding RNAs, particularly long non-coding RNAs (lncRNAs). These RNA molecules, exceeding 200 nucleotides in length and lacking protein-coding capacity, have emerged as pivotal players in HCC, influencing its initiation, progression, invasion, and metastasis by modulating gene expression at epigenetic, transcriptional, and post-transcriptional levels [16]. This application note details the molecular signatures, functional mechanisms, and experimental protocols for six key lncRNA candidates—HULC, UCA1, LINC00152, GAS5, MALAT1, and HOTAIR—framed within an integrative machine learning approach for advanced HCC diagnostics and therapeutic development.
Molecular Mechanisms and Pathogenic Significance
The oncogenic and tumor-suppressive lncRNAs characterized here contribute to HCC progression through diverse and overlapping signaling pathways.
Oncogenic lncRNAs and Their Pathways
HULC : The Highly Upregulated in Liver Cancer (HULC) lncRNA is stabilized in the HCC cellular environment and promotes tumor growth by elevating cyclooxygenase-2 (COX-2) protein levels. This stabilization is achieved through enhanced expression of ubiquitin-specific peptidase 22 (USP22), which removes conjugated polyubiquitin chains from COX-2, thereby inhibiting its proteasomal degradation [18]. HULC also functions as a competing endogenous RNA (ceRNA), sequestering miRNAs like miRNA-372 and reducing their inhibitory effect on target genes such as PRKACB, ultimately activating autophagy and promoting hepatoma cell proliferation [16].
UCA1 : Upregulated by the Hepatitis B virus X (HBx) protein, UCA1 promotes cell growth by facilitating the G1/S transition. It physically associates with the histone methyltransferase EZH2 (a component of the Polycomb Repressive Complex 2), which subsequently suppresses the tumor suppressor p27Kip1 through histone H3 lysine 27 trimethylation (H3K27me3) on the p27Kip1 promoter. This HBx-UCA1/EZH2-p27Kip1 axis is a crucial signaling pathway in hepatocarcinogenesis [19].
MALAT1 : Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) acts as a proto-oncogene by upregulating the splicing factor SRSF1. This modulation leads to the production of anti-apoptotic splicing isoforms and activates the mTOR pathway via alternative splicing of S6K1, driving cellular transformation [20]. Furthermore, MALAT1 contributes to Wnt pathway activation, reinforcing its oncogenic potential [20].
HOTAIR : HOX Transcript Antisense RNA (HOTAIR) functions as a transcriptional modulator by recruiting two distinct chromatin-modifying complexes: the Polycomb Repressive Complex 2 (PRC2) and the LSD1/CoREST/REST complex. This coordinated action leads to the trimethylation of histone H3 on lysine 27 (H3K27me3) and the demethylation of histone H3 on lysine 4 (H3K4me2), resulting in the silencing of tumor suppressor genes. Its overexpression is strongly associated with metastasis, recurrence, and poor prognosis [21].
LINC00152 : This lncRNA promotes cell proliferation and tumor growth by cis-regulating the EpCAM promoter and activating the mTOR signaling pathway. Its promoter region is frequently hypomethylated in HCC, leading to its significant upregulation in tumor tissues [22].
Tumor-Suppressive lncRNA
GAS5 : In contrast to the oncogenic lncRNAs, Growth Arrest-Specific 5 (GAS5) acts as a tumor suppressor. It functions as a molecular sponge for miR-144-5p, thereby relieving the microRNA's repression of its target, Activating Transcription Factor 2 (ATF2). The GAS5/miR-144-5p/ATF2 axis enhances the radiosensitivity of HCC cells, and lower levels of GAS5 are found in radiation-resistant tissues [23].
Table 1: Core Functional Mechanisms of Key lncRNAs in HCC
| lncRNA | Expression in HCC | Primary Functional Mechanism | Key Interacting Molecules/Pathways |
|---|---|---|---|
| HULC | Upregulated [18] | Protein stabilization; ceRNA activity | USP22, COX-2, miR-372, PRKACB, SPHK1 [18] [16] |
| UCA1 | Upregulated (HBx-associated) [19] | Epigenetic silencing | EZH2, p27Kip1, CDK2 [19] |
| MALAT1 | Upregulated [20] | Splicing regulation; Pathway activation | SRSF1, mTOR, Wnt/β-catenin [20] |
| HOTAIR | Upregulated [21] | Chromatin remodeling | PRC2 (EZH2, SUZ12), LSD1 [21] |
| LINC00152 | Upregulated [22] | Transcriptional activation; Signaling pathway | EpCAM, mTOR [22] |
| GAS5 | Downregulated [23] | miRNA sponging | miR-144-5p, ATF2 [23] |
Table 2: Clinical Correlations of Key lncRNAs in HCC
| lncRNA | Correlation with Clinicopathological Features | Prognostic/Diagnostic Value |
|---|---|---|
| HULC | Positively correlated with Edmondson grade and HBV infection [16] | Potential plasma biomarker for HCC diagnosis [16] |
| UCA1 | Significant association with HBx presence in HCC tissues (P=0.028) [19] | Potential biomarker for HBx-driven hepatocarcinogenesis [19] |
| MALAT1 | Promotes tumor progression [21] | Potential biomarker for predicting HCC recurrence [21] |
| HOTAIR | Associated with lymph node metastasis, larger tumor size, and recurrence [21] | Powerful predictor of metastasis and survival [21] |
| LINC00152 | Significant correlation with tumor size (P=0.005) and Edmondson grade (P=0.002) [22] | Novel index for clinical diagnosis; stable in plasma/exosomes [22] |
| GAS5 | Lower levels in radiation-resistant HCC tissues [23] | Biomarker for predicting radiosensitivity and treatment response [23] |
Experimental Protocols for lncRNA Functional Analysis
Protocol 1: lncRNA Quantification and Validation
Objective: To accurately quantify lncRNA expression levels in HCC tissue and plasma samples. Reagents: TRI Reagent (Sigma), MirVana RNA Isolation Kit, PrimerScript RT Enzyme Mix I (TaKaRa), SYBR Premix Ex Taq II (TaKaRa), custom lncRNA-specific primers. Equipment: NanoDrop 2000 Spectrophotometer, GeneAmp PCR System 9700, LightCycler 480 II Real-time PCR Instrument. Procedure:
- RNA Extraction: Homogenize 30 mg frozen tissue or 200 μL plasma in TRI Reagent. Extract total RNA using the MirVana kit per manufacturer's protocol.
- RNA Quality Control: Determine RNA concentration and purity using NanoDrop (A260/A280 ratio ~2.0 is acceptable).
- Reverse Transcription (RT): Assemble 10 μL RT reactions containing 0.5 μg RNA, PrimerScript Buffer, oligo dT, random 6 mers, and PrimerScript RT Enzyme Mix I. Incubate: 37°C for 15 min, 85°C for 5 sec [17].
- Quantitative PCR (qPCR): Prepare 10 μL reactions with 1 μL cDNA, SYBR Green I Master, and lncRNA-specific primers. Run in triplicate on a LightCycler 480 II: 95°C for 10 min; 40 cycles of 95°C for 10 sec, 60°C for 30 sec [17].
- Data Analysis: Calculate relative expression using the 2^(-ΔΔCt) method with GAPDH or U6 as endogenous controls.
Protocol 2: Functional Characterization via Knockdown/Gain-of-Function
Objective: To determine the oncogenic or tumor-suppressive functions of lncRNAs through modulation of their expression. Reagents: Lipofectamine 3000, pcDNA3.1 overexpression vectors, small interfering RNAs (siRNAs), puromycin. Equipment: CO2 incubator, flow cytometer, fluorescent microscope. Procedure: A. Gene Modulation: 1. Overexpression: Clone full-length lncRNA into pcDNA3.1. Transfect HCC cells (e.g., HepG2, Huh7) using Lipofectamine 3000 [20]. 2. Knockdown: Transfert cells with lncRNA-specific siRNAs (e.g., 50 nM final concentration) using Lipofectamine 3000 [23]. For stable knockdown, use lentiviral shRNA vectors with puromycin selection (2 μg/mL for 96 hours) [20]. B. Functional Assays: 1. Proliferation Analysis: - CCK-8 Assay: Seed transfected cells in 96-well plates (2×10³ cells/well). Measure absorbance at 490nm at 24, 48, 72, and 96h post-seeding [22]. - Colony Formation: Seed 500-1000 transfected cells in 6-well plates. Culture for 10-14 days, fix with glutaraldehyde, and stain with 1% methylene blue. Count colonies [20] [19]. 2. Apoptosis Assay: 48h post-transfection, treat cells with pro-apoptotic agents if needed. Stain with Annexin V-FITC and PI. Analyze by flow cytometry [19]. 3. Cell Cycle Analysis: Fix cells in 70% ethanol, treat with RNase A, stain with propidium iodide, and analyze DNA content by flow cytometry [19]. 4. In Vivo Tumorigenesis: Subcutaneously inject 5×10^6 stably transfected HCC cells into flanks of 4-6 week-old BALB/C nude mice. Monitor tumor growth for 4-6 weeks [22].
Protocol 3: Mechanism of Action Studies
Objective: To identify molecular interactions and downstream pathways of target lncRNAs. Reagents: RIPA buffer, primary antibodies, Protein A/G beads, biotin-labeled lncRNA probes. Procedure:
- RNA-Protein Interaction:
- RNA Immunoprecipitation (RIP): Lyse cells in RIPA buffer. Incubate lysate with antibodies against target protein (e.g., EZH2) or control IgG. Precipitate with Protein A/G beads. Extract RNA from precipitates and analyze by qRT-PCR [23].
- RNA Pull-Down: Transcribe biotin-labeled lncRNA in vitro. Incubate with cell lysates. Capture RNA-protein complexes with streptavidin beads. Elute and identify bound proteins by western blot or mass spectrometry [23].
- Pathway Analysis: After lncRNA modulation, analyze key signaling pathways by western blotting for phosphorylated/ total proteins (e.g., p-mTOR/mTOR, COX-2) [18] [22].
Visualizing Molecular Relationships and Workflows
HCC-Associated lncRNA Molecular Relationships
Experimental Workflow for lncRNA Biomarker Development
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Research Reagents for lncRNA HCC Research
| Reagent/Catalog | Primary Application | Experimental Function |
|---|---|---|
| TRI Reagent (Sigma) | RNA Extraction | Simultaneous isolation of high-quality RNA, DNA, and proteins from tissue/cell samples [17]. |
| mirVana RNA Isolation Kit | RNA Purification | Specialized column-based isolation of total RNA, enriched for small RNAs including lncRNAs [17]. |
| Lipofectamine 3000 | Cell Transfection | Lipid-based reagent for efficient delivery of nucleic acids (siRNA, plasmids) into mammalian cells [23]. |
| SYBR Green Master Mix | qRT-PCR | Fluorescent dye for detection and quantification of PCR products in real-time [23]. |
| Annexin V-FITC/PI Kit | Apoptosis Assay | Flow cytometry-based detection of early and late apoptotic cell populations [19]. |
| Cell Counting Kit-8 (CCK-8) | Proliferation Assay | Colorimetric assay for sensitive quantification of viable cells in proliferation/cytotoxicity studies [22]. |
| Puromycin Dihydrochloride | Stable Cell Selection | Antibiotic for selection of mammalian cells stably transfected with puromycin resistance genes [20]. |
| RIPA Lysis Buffer | Protein Extraction | Efficient extraction of total cellular protein for downstream western blotting and immunoprecipitation [23]. |
| 2-Acetylbenzoic acid | 2-Acetylbenzoic acid, CAS:577-56-0, MF:C9H8O3, MW:164.16 g/mol | Chemical Reagent |
| Sphondin | Sphondin, CAS:483-66-9, MF:C12H8O4, MW:216.19 g/mol | Chemical Reagent |
Integration with Machine Learning Frameworks
The transition from bench to bedside for lncRNA biomarkers requires robust computational integration. Machine learning (ML) algorithms can efficiently analyze complex RNA expression patterns from high-throughput sequencing data to identify novel biomarker signatures with diagnostic, prognostic, and predictive utility [9]. Support Vector Machines (SVMs) and neural networks have been successfully trained using circulating RNA data to differentiate between benign and malignant liver diseases [9]. For HCC biomarker development, ML pipelines typically integrate:
- Feature Selection: Identification of the most discriminative lncRNAs from transcriptomic datasets.
- Model Training: Utilizing algorithms like Random Forest and XGBoost, which have proven effective in identifying critical genes in cancer pathogenesis [9].
- Multi-Omics Integration: Combining lncRNA expression profiles with genomic, epigenomic, and clinical data to generate comprehensive diagnostic signatures that enhance early detection rates and minimize false positives [9].
This integrated approach facilitates the development of clinically viable lncRNA biomarker panels that can transform HCC management through improved early detection, accurate prognosis prediction, and personalized treatment strategies.
Long non-coding RNAs (lncRNAs), defined as transcripts longer than 200 nucleotides that do not code for proteins, have emerged as promising biomarkers for liquid biopsy due to their stability in biofluids and deep involvement in cancer pathogenesis [24]. Their utility is particularly pronounced in hepatocellular carcinoma (HCC), where the need for non-invasive diagnostic tools is critical given the risks and limitations associated with traditional liver biopsies [25] [26]. LncRNAs are remarkably stable in circulation through their packaging into membrane-bound vesicles like exosomes or through complex formation with RNA-binding proteins such as Argonaute 2 (AGO2) and lipoproteins [24]. This stability, combined with their disease-specific expression patterns, makes them ideal candidates for developing sensitive and specific diagnostic assays.
The integration of lncRNA biomarkers with machine learning (ML) algorithms represents a transformative approach for HCC diagnosis, moving beyond single-marker thresholds to multi-analyte predictive models. This integration leverages the strengths of both molecular biology and computational science to achieve superior diagnostic performance [27] [14]. This Application Note details the experimental protocols for lncRNA handling and analysis, contextualized within a framework for machine learning integration in HCC diagnostics.
Stability and Origin of Cell-Free lncRNAs
Understanding the mechanisms that confer stability to cell-free lncRNAs is fundamental to developing robust liquid biopsy assays. The following table summarizes the primary forms and protective mechanisms of circulating lncRNAs.
Table 1: Forms and Stability Mechanisms of Cell-Free lncRNAs
| Form | Protective Mechanism | Key Characteristics | Implications for Liquid Biopsy |
|---|---|---|---|
| Exosomes & Extracellular Vesicles (EVs) | Encapsulation within lipid bilayer membranes [24] [28]. | Double-layered membrane shields contents from RNases; carries tumor-specific molecular markers (e.g., EpCAM) [28]. | Provides high stability; enables tumor origin specificity via surface marker isolation. |
| Protein Complexes | Binding to RNA-binding proteins like Argonaute 2 (AGO2) [24]. | Protection without membrane encapsulation; mechanism distinct from vesicular packaging. | Contributes to the overall pool of stable cell-free lncRNAs detectable in plasma. |
| Lipoprotein Complexes | Association with High-Density Lipoproteins (HDLs) [24]. | Protection without membrane encapsulation; alternative stability mechanism. | Another source of stable lncRNA for detection, complementing vesicular and protein-bound fractions. |
The origin of these lncRNAs is equally important. Tumor-released exosomes faithfully reflect the molecular signature of their parental cells. For instance, exosomes bearing epithelial cell adhesion molecule (EpCAM) are significantly elevated in cancer patients and contain lncRNAs that show significant concordance with tumor tissue expressions, making them a highly specific substrate for analysis [28].
Experimental Protocols for lncRNA Analysis
Plasma Collection and Exosome Isolation
Protocol: Plasma Exosome Isolation via Precipitation
- Blood Collection and Pre-processing: Collect peripheral blood using heparin or EDTA tubes. Centrifuge at 3,000 × g for 15 minutes at 4°C to pellet cells and debris [28].
- Exosome Precipitation: Transfer the clarified plasma to a fresh tube. Add the recommended volume of exosome precipitation solution (e.g., ExoQuick, SBI). Mix thoroughly by inverting and incubate at 4°C for 30-60 minutes [28].
- Exosome Pellet Formation: Centrifuge the mixture at 3,000 × g for 10-30 minutes. A beige or white pellet should be visible at the bottom of the tube. Carefully aspirate the supernatant without disturbing the pellet [28].
- Resuspension: Resuspend the exosome pellet in a suitable buffer (e.g., nuclease-free PBS or RNAse-free water) for downstream applications. Isolated exosomes can be stored at -80°C [28].
Protocol: Immunoaffinity Capture of Tumor-Specific Exosomes
For enhanced specificity, exosomes from tumor cells can be isolated using antibodies against surface markers like EpCAM [28].
- Bead Preparation: Dispense EpCAM-coated magnetic beads into a tube.
- Sample Incubation: Add pre-cleared plasma and incubation buffer to the beads. Incubate with gentle mixing for at least 30 minutes to allow exosomes to bind.
- Washing: Place the tube on a magnetic stand, discard the supernatant, and wash the beads with buffer to remove non-specifically bound material.
- Elution (Optional): For some applications, captured exosomes can be eluted using a low-pH or detergent-based elution buffer. Alternatively, lysis buffer can be added directly to the beads for RNA extraction [28].
Validation: Isolated exosomes should be characterized for size and morphology using Transmission Electron Microscopy (TEM) and nanoparticle tracking analysis (NanoFCM). The presence of exosomal markers (e.g., CD63, CD81) and the specific capture marker (e.g., EpCAM) can be confirmed by western blot [28].
RNA Extraction and Quality Control
Protocol: Total RNA Isolation from Plasma or Exosomes
- Lysis: Mix the plasma or resuspended exosome sample with a lysis buffer containing a denaturing guanidine-isothiocyanate solution to inactivate RNases.
- RNA Binding: Pass the lysate through a silica-based membrane column. RNA binds to the membrane under high-salt conditions, while contaminants are washed away.
- Washing: Perform two wash steps using ethanol-containing buffers to remove salts and other impurities.
- Elution: Elute the pure RNA in a small volume of nuclease-free water. Recommended kits include the miRNeasy Mini Kit (Qiagen) or Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit (Norgen Biotek) [27] [25].
Quality Control: Quantify RNA concentration using a fluorometer (e.g., Qubit with RNA HS Assay Kit). Due to low yields, quality assessment via Bioanalyzer may not be feasible; therefore, the integrity of the reverse transcription and qPCR reaction serves as a functional quality check [14].
lncRNA Quantification by qRT-PCR
Protocol: Reverse Transcription and Quantitative PCR
- cDNA Synthesis: Reverse transcribe purified RNA using a High-Capacity cDNA Reverse Transcription Kit. Include genomic DNA removal steps (e.g., DNase I treatment) [27] [25].
- qPCR Setup: Perform qPCR reactions using Power SYBR Green Master Mix or TaqMan assays on a real-time PCR system (e.g., ViiA 7 or StepOne Plus). Each reaction should be performed in triplicate.
- Data Analysis: Use the comparative Ct (ΔΔCt) method for relative quantification. Normalize lncRNA expression to a stable endogenous control (e.g., GAPDH, β-actin, or SNORD72 for plasma RNA) [27] [25].
Table 2: Example Primers for HCC-Associated lncRNAs
| lncRNA | Sense Primer (5' to 3') | Antisense Primer (5' to 3') | Application Context |
|---|---|---|---|
| LINC00152 | GACTGGATGGTCGCTTT | CCCAGGAACTGTGCTGTGAA | Diagnostic panel for HCC [27] |
| UCA1 | TGCACCGACCCGAAACT | CAAGTGTGACCAGGGACTGC | Diagnostic panel for HCC [27] |
| GAS5 | TCCCAGCCTCAGACTCAACA | TCGTGTCC | Diagnostic & prognostic panel for HCC [27] |
| LINC00853 | AAAGGCTAGGCGATCCCACA | ACTCCCTAGCTTGGCTCTCCT | Diagnostic panel for HCC [27] |
| RP11-731F5.2 | Information in source [25] | Information in source [25] | Biomarker for HCC risk in CHC patients [25] |
Integration with Machine Learning for HCC Diagnosis
The true power of lncRNA signatures is unlocked when multiple markers are combined using machine learning models, moving beyond univariate analysis.
Data Preparation and Feature Engineering
The first step is to create a structured data matrix for model training.
- Features: Normalized expression values (ΔCt or RQ) of a panel of lncRNAs (e.g., from Table 2), combined with standard clinical variables (e.g., AFP, ALT, AST, age, cirrhosis status) [27] [14].
- Outcome Label: The diagnostic status (HCC vs. non-HCC) or prognostic outcome (e.g., high-risk vs. low-risk recurrence) for each patient.
Table 3: Machine Learning Models for lncRNA-Based HCC Diagnosis
| Model | Key Characteristics | Reported Performance in HCC Context |
|---|---|---|
| Light Gradient Boosting Machine (LGBM) | A highly efficient gradient-boosting framework that uses tree-based algorithms. | Achieved 98.75% accuracy in diagnosing HCC using an 8-RNA signature panel [14]. |
| Random Survival Forest (RSF) | An ensemble learning method for survival data, effective for prognostic risk stratification. | Used to develop a 6-gene prognostic risk score for HCC with high accuracy (C-index) [29]. |
| Support Vector Machine (SVM) | Finds an optimal hyperplane to separate different classes in a high-dimensional space. | One of multiple algorithms evaluated in a 10-model framework for prognostic modeling [29]. |
| LASSO Cox Regression | Performs both variable selection and regularization to enhance prediction accuracy. | Commonly used for selecting the most relevant features in high-dimensional genomic data [15] [30]. |
Model Training and Workflow
The general workflow for building an HCC diagnostic model involves feature selection, model training, and validation.
Figure 1: Machine learning integration workflow for lncRNA-based HCC diagnosis.
The Scientist's Toolkit: Key Research Reagents
Table 4: Essential Reagents and Kits for lncRNA Liquid Biopsy Research
| Reagent / Kit | Function | Example Product / Vendor |
|---|---|---|
| Exosome Isolation Kit | Precipitates total exosomes from plasma/serum. | ExoQuick (SBI) [28] |
| Immunomagnetic Beads | Isulates tumor-specific exosomes via surface markers. | EpCAM-coated magnetic beads [28] |
| RNA Extraction Kit | Purifies high-quality total RNA from plasma/exosomes. | miRNeasy Mini Kit (Qiagen) [27] [25] |
| cDNA Synthesis Kit | Reverse transcribes RNA into stable cDNA. | High-Capacity cDNA Kit (Thermo Fisher) [25] |
| SYBR Green Master Mix | For fluorescence-based qPCR quantification. | Power SYBR Green (Thermo Fisher) [27] |
| NanoParticle Analyzer | Characterizes exosome size distribution and concentration. | NanoFCM N30E [28] |
| Finasteride-d9 | Finasteride-d9 | High Purity Stable Isotope | RUO | Finasteride-d9 internal standard for accurate LC-MS/MS quantification. For Research Use Only. Not for human or veterinary diagnostic or therapeutic use. |
| Pamidronic Acid | Pamidronic Acid|High-Purity Research Reagent | High-purity Pamidronic Acid, a potent bisphosphonate for bone metabolism and oncology research. This product is For Research Use Only (RUO). Not for human or veterinary use. |
The protocols outlined herein provide a robust framework for leveraging plasma and exosomal lncRNAs as non-invasive biomarkers for HCC. The critical steps—careful sample collection, specific exosome isolation, rigorous RNA quantification, and data integration via machine learning—are paramount for success. Future advancements will rely on the standardization of these protocols across laboratories and the validation of lncRNA signatures in large, multi-center prospective cohorts. The convergence of liquid biopsy technology and machine learning analytics holds the definitive promise of transforming HCC management, enabling earlier detection, accurate prognosis, and personalized therapeutic strategies.
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related mortality globally, with prognosis heavily dependent on early detection. For decades, alpha-fetoprotein (AFP) has been the most widely used serological biomarker for HCC surveillance. However, its diagnostic performance is suboptimal, particularly for early-stage tumors, with sensitivity reported as low as 50-70% [31] [32]. This limitation has spurred the investigation of novel biomarkers, notably long non-coding RNAs (lncRNAs), which show deregulated expression in hepatocarcinogenesis. The integration of these RNA biomarkers with artificial intelligence (AI) analysis frameworks represents a transformative approach for improving HCC diagnosis, offering significant enhancements in both sensitivity and specificity compared to traditional AFP testing.
Performance Comparison: Traditional vs. Novel Biomarker Approaches
The quantitative superiority of lncRNA and AI-driven approaches over AFP is evident across multiple clinical studies. The table below summarizes key performance metrics from recent research.
Table 1: Performance Comparison of HCC Diagnostic Approaches
| Biomarker / Approach | Sensitivity (%) | Specificity (%) | AUC/Other Metrics | Study Focus |
|---|---|---|---|---|
| Alpha-fetoprotein (AFP) | 50-70 [31] | - | - | MRD detection post-treatment [31] |
| AFP (Early HCC) | Lower than AI model [32] | Lower than AI model [32] | Suboptimal for early-stage [32] | Early-stage HCC detection |
| lncRNA Panel (LINC00152, LINC00853, UCA1, GAS5) + ML | 100 [6] | 97 [6] | - | HCC diagnosis vs. controls |
| Blood-based AI Model (Routine tests) | 80 [32] | 81 [32] | AUROC: 0.894 [32] | Early-stage detection in CLD |
| Plasma lncRNA HULC | - | - | - | HCC risk in CHC patients [33] [25] |
| Machine Learning (RF Model for HBV-cACLD) | 80.8 [34] | - | AUC: 0.979 [34] | HCC risk prediction |
MRD: Minimal Residual Disease; CLD: Chronic Liver Disease; CHC: Chronic Hepatitis C; HBV-cACLD: Hepatitis B Virus-related compensated Advanced Chronic Liver Disease; RF: Random Forest.
The data consistently demonstrates that multi-analyte panels analyzed via machine learning outperform the single-marker AFP test. The AI model using standard blood tests achieved an 80% sensitivity for early-stage HCC, a significant improvement over AFP alone [32]. Remarkably, a model integrating a four-lncRNA expression panel with clinical parameters achieved 100% sensitivity and 97% specificity [6].
Experimental Protocols for lncRNA Biomarker Research
Protocol 1: Liquid Biopsy for Plasma lncRNA Analysis
This protocol outlines the process for quantifying circulating lncRNAs from patient plasma, a key method for non-invasive biomarker discovery [33] [6] [25].
1. Sample Collection and Processing:
- Collect peripheral blood into EDTA or citrate tubes.
- Centrifuge at 704 × g (RCF) for 10 minutes at 4°C to separate plasma from cellular components.
- Carefully aliquot the supernatant plasma and store at -70°C until RNA extraction.
2. RNA Isolation:
- Use a commercial Plasma/Serum Circulating and Exosomal RNA Purification Kit.
- Process 500 μL of plasma per the manufacturer's protocol.
- Treat the isolated RNA with Turbo DNase to remove genomic DNA contamination.
3. cDNA Synthesis:
- Use a High-Capacity cDNA Reverse Transcription Kit.
- Perform reverse transcription using a thermal cycler with the following conditions: 10 minutes at 25°C, 120 minutes at 37°C, and 5 minutes at 85°C.
4. Quantitative Real-Time PCR (qRT-PCR):
- Use Power SYBR Green PCR Master Mix on a real-time PCR system.
- Prepare reactions in triplicate, including no-template controls.
- Use the following cycling conditions: initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec and 62°C for 1 min.
- Use β-actin or GAPDH as an internal reference gene for normalization.
- Confirm reaction specificity by performing a dissociation melting curve analysis.
5. Data Analysis:
- Calculate relative expression levels using the 2^(-ΔΔCt) method [33] [6].
- Perform statistical analysis and generate Receiver Operating Characteristic (ROC) curves to evaluate the diagnostic power of individual lncRNAs.
Protocol 2: Developing a Machine Learning Diagnostic Model
This protocol describes the workflow for building a machine learning model to integrate lncRNA data with clinical features for superior HCC diagnosis [34] [6].
1. Data Collection and Cohort Definition:
- Case Group: Recruit patients with HCC diagnosed via histopathology or non-invasive imaging criteria (e.g., LI-RADS).
- Control Group: Recruit age-matched controls, including healthy individuals and patients with chronic liver disease (e.g., chronic hepatitis C) but without HCC.
- Collect relevant clinical and laboratory data (e.g., ALT, AST, AFP, bilirubin, albumin).
2. Feature Selection:
- To avoid overfitting and identify the most predictive variables, apply feature selection algorithms on the training cohort only.
- Least Absolute Shrinkage and Selection Operator (LASSO): Applies L1 regularization to shrink less important feature coefficients to zero.
- Random Forest (RF): Ranks feature importance based on the mean decrease in Gini impurity.
- Support Vector Machine (SVM): Ranks features using average rank (AvgRank). Select key predictors that are identified by multiple methods.
3. Machine Learning Model Construction and Training:
- Randomly split the dataset into a training cohort (e.g., 70-80%) and a validation cohort (e.g., 20-30%).
- Construct multiple models on the training set using selected features. Common algorithms include:
- Random Forest: An ensemble of decision trees.
- Support Vector Machine (SVM): Can use linear or radial basis function (RBF) kernels.
- Logistic Regression: Often with L2 regularization.
- Extreme Gradient Boosting (XGBoost): An efficient implementation of gradient boosting.
- Optimize model hyperparameters via grid search or cross-validation.
4. Model Validation and Interpretation:
- Evaluate the final model's performance on the held-out validation cohort using metrics such as Accuracy, Sensitivity, Specificity, and Area Under the ROC Curve (AUC).
- Employ model interpretation tools like SHapley Additive exPlanations (SHAP) to quantify the contribution of each feature to the model's predictions, enhancing clinical translatability [34].
Workflow Visualization
lncRNA Biomarker Discovery & Validation
AI Integration for HCC Diagnosis
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Kits for lncRNA Biomarker Research
| Item | Function/Application | Example Product(s) |
|---|---|---|
| Plasma/Serum RNA Kit | Isolation of high-quality circulating and exosomal RNA from plasma/serum. | Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit (Norgen Biotek) [33] [25] |
| DNase Treatment Kit | Removal of genomic DNA contamination from RNA samples to ensure pure template. | Turbo DNase (Life Technologies) [33] [25] |
| cDNA Synthesis Kit | Reverse transcription of RNA into stable cDNA for downstream qPCR applications. | High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher) [33] [25]; RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) [6] |
| qRT-PCR Master Mix | Sensitive and specific detection and quantification of lncRNA targets via SYBR Green chemistry. | Power SYBR Green PCR Master Mix (Thermo Fisher) [33] [6] [25]; PowerTrack SYBR Green Master Mix (Applied Biosystems) [6] |
| Specific lncRNA Primers | Target-specific amplification of lncRNAs of interest (e.g., HULC, LINC00152, GAS5). | Custom-designed primers from suppliers like Thermo Fisher Scientific [6] |
| Methyl 3,4-dimethoxycinnamate | Methyl 3,4-dimethoxycinnamate, CAS:5396-64-5, MF:C12H14O4, MW:222.24 g/mol | Chemical Reagent |
| Ansatrienin A | Mycotrienin I|Potent Inhibitor of Bone Resorption | Mycotrienin I is a potent ansamycin antibiotic that inhibits osteoclastic bone resorption. For Research Use Only. Not for human or veterinary use. |
The integration of lncRNA biomarkers with machine learning analytics marks a significant leap forward in the quest for precision oncology in HCC. The evidence confirms that this approach consistently surpasses the diagnostic performance of the traditional AFP test, offering markedly improved sensitivity and specificity for early detection. While challenges in standardization and clinical validation remain, the protocols and tools outlined herein provide a clear roadmap for researchers and drug development professionals to advance this promising field, ultimately contributing to improved patient outcomes through earlier and more accurate diagnosis.
Building Diagnostic Power: Machine Learning Algorithms and Workflows for lncRNA Signature Development
Within the framework of advancing the machine learning integration of long non-coding RNA (lncRNA) biomarkers for Hepatocellular Carcinoma (HCC) diagnosis, the acquisition and rigorous preprocessing of high-quality genomic data constitutes a critical foundational step. The accuracy and reliability of subsequent predictive models are fundamentally dependent on the integrity of the underlying data. This protocol details comprehensive methodologies for sourcing lncRNA expression data from two premier public repositories, The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO), and preparing it for downstream machine learning applications. The procedures outlined herein are designed to equip researchers, scientists, and drug development professionals with a standardized workflow to construct robust, analysis-ready datasets, thereby facilitating the discovery and validation of novel lncRNA diagnostic signatures for HCC.
Data Sourcing from Primary Repositories
Table 1: Primary Data Repositories for lncRNA Expression Data
| Repository | Data Type | Key HCC Datasets | Primary Access Method |
|---|---|---|---|
| The Cancer Genome Atlas (TCGA) | Clinical data, RNA-seq (lncRNA, mRNA), miRNA, DNA methylation, somatic mutations [35] [36] | TCGA-LIHC (Liver Hepatocellular Carcinoma) [37] [38] | GDC Data Portal, TCGAbiolinks R package [35] [36] |
| Gene Expression Omnibus (GEO) | Curated gene expression datasets from microarray and NGS studies [39] [40] | GSE14520, GSE57555, GSE19665, among others [40] [41] | GEO2R, manual download from NCBI [41] |
Accessing Data from The Cancer Genome Atlas (TCGA)
TCGA provides a comprehensive, multi-omics view of over 30 cancer types, including HCC (project code: TCGA-LIHC). Data access is primarily facilitated through the Genomic Data Commons (GDC) Data Portal and programmatic interfaces [35].
Protocol 2.1: Downloading TCGA Data via the GDC Data Portal
- Navigate to the Portal: Access the GDC Data Portal at https://portal.gdc.cancer.gov/.
- Select the HCC Project:
- Click on "Projects" in the top navigation.
- Within the "Programs" filter, select "TCGA".
- Locate and select "TCGA-LIHC" from the resulting list.
- Build a Cohort (Optional): Use the "Cohort Builder" to refine cases based on clinical or molecular characteristics (e.g., select only female subjects or specific tumor stages).
- Access the Repository for Files: Navigate to the "Repository" tab to filter and select specific files for download.
- Apply File Filters:
- Download Files:
- Add the desired files to the cart.
- Download a "Manifest" file for use with the GDC Data Transfer Tool (recommended for large datasets).
- Alternatively, for datasets under 5 GB, use the "Download Cart" option directly.
- Ensure you also download the associated clinical and biospecimen metadata files.
Protocol 2.2: Programmatic Access using R and TCGAbiolinks
The following R code provides a robust method for querying and downloading TCGA data directly into an analysis environment.
Code 1: Querying, downloading, and preparing TCGA-LIHC data using R.
It is crucial to distinguish between Harmonized data (aligned to the GRCh38 reference genome and processed through standardized GDC pipelines) and Legacy data (the original data generated by TCGA centers). For new analyses, the use of harmonized data is strongly recommended to ensure consistency [35].
Accessing Data from the Gene Expression Omnibus (GEO)
GEO is a public repository that archives and freely distributes high-throughput gene expression and other functional genomics datasets submitted by the research community [40] [41].
Protocol 2.3: Identifying and Downloading HCC-relevant Data from GEO
- Search and Identify Datasets: Use the GEO DataSet browser (https://www.ncbi.nlm.nih.gov/gds/) with keywords such as "Hepatocellular Carcinoma," "HCC," "lncRNA," and "Homo sapiens".
- Review Dataset Landing Page: Carefully examine the dataset's description (GSE page) to ensure it includes HCC and normal tissue samples and utilizes a platform suitable for lncRNA detection.
- Download Data:
- Processed Data: Download the series matrix file (
*_series_matrix.txt.gz) containing the normalized expression values and sample metadata. - Raw Data: For re-analysis, download the raw data files (e.g., .CEL files for Affymetrix platforms) from the "Supplementary files" section.
- Processed Data: Download the series matrix file (
- Utilize GEO2R for Quick Analysis: GEO2R is an interactive web tool that allows users to compare groups of samples to identify differentially expressed genes directly within the browser. While useful for initial exploration, it is not a substitute for a full, reproducible bioinformatics pipeline for machine learning projects [41].
Data Preprocessing and Curation
Raw genomic data must be processed and normalized to create a reliable dataset for machine learning model training. The workflow below outlines the key stages.
Diagram 1: Data preprocessing workflow for lncRNA expression data.
Quality Control and Filtering
The initial step involves assessing data quality and removing uninformative genes.
- Quality Metrics: For RNA-seq data, metrics include total read count, alignment rate, and genomic distribution of reads. For microarray data, inspection of log-intensity distributions and RNA degradation plots is standard.
- Filtering Low-Expressed Genes: Genes with very low counts across most samples can introduce noise. A common filter is to retain only lncRNAs and mRNAs with a count per million (CPM) above a threshold (e.g., 1 CPM) in a minimum number of samples (e.g., the size of the smallest group of samples) [37] [38]. This step reduces the feature space and improves the power of subsequent statistical tests.
Normalization and Batch Effect Correction
Normalization adjusts for technical variations (e.g., sequencing depth, library preparation) to make expression levels comparable between samples.
Protocol 3.1: Normalization of RNA-seq Count Data
For downstream analyses like differential expression and machine learning, it is essential to use normalized data. The edgeR and DESeq2 packages in R are widely used for this purpose.
Code 2: Normalizing RNA-seq count data using the edgeR package in R.
Batch effects are technical sources of variation arising from processing samples in different batches, dates, or platforms. They can severely confound machine learning models. The sva R package contains the ComBat function, which is a commonly used tool for adjusting for batch effects in high-dimensional genomic data [36].
Integration with Machine Learning Workflows
Once preprocessed, the data can be formatted for machine learning tasks, such as building a diagnostic signature.
Table 2: Key lncRNA Biomarkers for HCC Diagnosis and Prognosis from Literature
| lncRNA Name | Expression in HCC | Potential Clinical Role | Reported Performance (AUC/Sensitivity/Specificity) | Source |
|---|---|---|---|---|
| 4-lncRNA Signature (AC108463.1, AF131217.1, CMB9-22P13.1, TMCC1-AS1) | Risk Score | Prognosis (Early Recurrence) | Combined with AFP & TNM improved predictive performance [37] | TCGA |
| CRNDE | Upregulated | Diagnosis | AUC: 0.701; Sens: 71.0%; Spec: 87.1% [40] | GEO, TCGA |
| LINC00152 | Upregulated | Diagnosis, Prognosis | Machine learning model combining 4 lncRNAs achieved 100% Sens, 97% Spec [6] | Patient Plasma |
| RP11-486O12.2, LINC01093, et al. | Dysregulated | Diagnosis | Random Forest/SVM model AUC: 0.992 [38] | TCGA |
Protocol 4.1: Constructing a Machine Learning-Ready Dataset
- Merge Data Matrices: Combine the normalized lncRNA expression matrix with relevant clinical variables (e.g., age, gender, AFP levels, TNM stage) into a single data frame.
- Define the Outcome Variable: Specify the target variable for the machine learning model (e.g.,
Sample_Typewith levels "Tumor" vs. "Normal" for diagnosis, orRecurrence_Statusfor prognosis). - Partition Data: Split the complete dataset into training (e.g., 70-80%) and testing (e.g., 20-30%) sets, ensuring stratified sampling to preserve the distribution of the outcome variable in both sets.
- Feature Selection: Apply machine learning-driven feature selection techniques to identify the most predictive lncRNAs. Common methods include:
- LASSO (Least Absolute Shrinkage and Selection Operator): Penalizes the absolute size of regression coefficients, effectively driving coefficients of non-informative features to zero [37] [38].
- Random Forest: Ranks features by their importance based on the decrease in model accuracy when the feature's values are permuted [37] [38].
- SVM-RFE (Support Vector Machine-Recursive Feature Elimination): Recursively removes features with the smallest weights and rebuilds the SVM model to find an optimal feature subset [37].
The final output is a clean, formatted table where rows are samples, columns are features (lncRNA expression levels and clinical variables), and one column is the designated outcome, ready for input into machine learning algorithms.
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions and Computational Tools
| Item / Tool Name | Function / Application | Relevant Context in HCC lncRNA Research |
|---|---|---|
| miRNeasy Kit (QIAGEN) | Isolation of total RNA (including lncRNAs) from tissues and biofluids. | Used for plasma RNA isolation in studies identifying circulating lncRNA biomarkers like LINC00152 and UCA1 [6]. |
| PowerTrack SYBR Green Master Mix | Sensitive detection and quantification of lncRNAs via qRT-PCR. | Validation of differentially expressed lncRNAs (e.g., CRNDE, LINC01419) identified from bioinformatics analysis [40] [6]. |
| TCGAbiolinks R Package | Programmatic access, integration, and analysis of TCGA data. | Downloading and preparing TCGA-LIHC data for identification of diagnostic lncRNA signatures [36] [38]. |
| TANRIC (The Atlas of non-coding RNA in Cancer) | Interactive open platform to explore lncRNA function and expression. | Used in cross-platform studies to explore the clinical relevance of identified lncRNA biomarker candidates [39] [42]. |
| DESeq2 / edgeR R Packages | Differential expression analysis of RNA-seq data. | Statistical identification of lncRNAs dysregulated in HCC compared to normal tissues [37] [38]. |
| Scikit-learn (Python Library) | Machine learning library for building predictive models. | Construction of a diagnostic model integrating lncRNA expression and clinical laboratory data [6]. |
| 6,7-Dihydroxy-4-coumarinylacetic acid | 6,7-Dihydroxy-4-coumarinylacetic acid, CAS:88404-14-2, MF:C11H8O6, MW:236.18 g/mol | Chemical Reagent |
| (S)-Venlafaxine | (S)-Venlafaxine|High-Purity SNRI for Research |
Within the broader scope of integrating machine learning with long non-coding RNA (lncRNA) biomarkers for Hepatocellular Carcinoma (HCC) diagnosis, the precise identification of critical molecular features from high-dimensional transcriptomic data represents a fundamental challenge. The selection of biologically relevant and non-redundant lncRNA signatures directly dictates the performance, interpretability, and clinical translatability of prognostic and diagnostic models. This Application Note details the established protocols for three dominant feature selection techniques—LASSO, Random Forest, and SVM-RFE—that have been rigorously validated for lncRNA biomarker discovery in HCC research. We provide a structured framework for their implementation, enabling researchers to systematically isolate the most informative lncRNAs from complex expression datasets.
Core Feature Selection Techniques: Principles and Applications
The following techniques are instrumental in refining vast lncRNA expression datasets into potent, minimal biomarker signatures.
Least Absolute Shrinkage and Selection Operator (LASSO) operates as a regularization technique that applies an L1 penalty to the regression coefficients. This penalty effectively shrinks less important coefficients to zero, thereby performing automatic variable selection. Its primary application in lncRNA research is for constructing parsimonious prognostic signatures, particularly in high-dimensional settings where the number of features (lncRNAs) vastly exceeds the number of observations (patients) [43] [15]. A notable application includes the development of a 25-lncRNA signature for predicting early recurrence in HCC, where LASSO was pivotal in distilling the final candidate lncRNAs from an initial pool of candidates [43].
Random Forest (RF) is an ensemble learning method that constructs multiple decision trees. Its feature importance metric, often based on the mean decrease in Gini impurity or accuracy, provides a robust measure for ranking lncRNAs. This method is highly effective for non-linear data and captures complex interactions between features, making it suitable for initial screening and prioritization of a larger set of lncRNAs [15] [38]. In one study, the top 30 lncRNAs ranked by Random Forest importance were selected for further analysis in building a 4-lncRNA prognostic signature [15].
Support Vector Machine-Recursive Feature Elimination (SVM-RFE) is a wrapper method that utilizes the weights of a Support Vector Machine model to rank features. It recursively removes the least important features (e.g., those with the smallest absolute weights) and rebuilds the model until an optimal feature subset is identified. SVM-RFE is widely used for identifying diagnostic lncRNA biomarkers, as it effectively finds features that maximize the separation between classes, such as HCC versus normal tissue [15] [44] [38].
Table 1: Comparative Analysis of Feature Selection Techniques for lncRNA Biomarker Discovery
| Technique | Mechanism | Primary Strength | Typical Application in HCC lncRNA Studies | Example Signature Outcome |
|---|---|---|---|---|
| LASSO (L1 Regularization) | Shrinks coefficients, zeroing out irrelevant features | Prevents overfitting; creates sparse, interpretable models | Prognostic signature development for survival/ recurrence [43] [15] | 25-lncRNA [43] and 4-lncRNA [15] early recurrence signatures |
| Random Forest | Ranks features by mean decrease in Gini/accuracy | Robust to outliers; captures complex, non-linear interactions | Initial feature screening and prioritization from a large candidate pool [15] [38] | Selection of top 30 features for downstream refinement [15] |
| SVM-RFE | Recursively eliminates features with smallest SVM weights | Maximizes separation between classes (e.g., Tumor vs. Normal) | Diagnostic biomarker identification [38] | 4-lncRNA diagnostic panel (RP11‑486O12.2, RP11‑863K10.7, LINC01093, RP11‑273G15.2) [38] |
Integrated Experimental Protocol for lncRNA Signature Development
This section outlines a standardized workflow for identifying and validating a prognostic lncRNA signature in HCC, integrating the feature selection techniques described above.
Data Acquisition and Preprocessing
- Data Source: Obtain lncRNA expression data (e.g., RNA-seq or microarray) and corresponding clinical data (e.g., disease-free survival, overall survival) from public repositories such as The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) project [43] [15] [38].
- Cohort Division: Randomly split the patient cohort into a training set (e.g., 50%) and a validation set (e.g., 50%). All subsequent feature selection and model building must occur exclusively within the training cohort [43] [15].
- Differential Expression Analysis: Identify differentially expressed lncRNAs (DElncs) between tumor and adjacent normal tissues in the training cohort using packages such as
DESeq2,edgeR, orlimmain R. Apply a false discovery rate (FDR) < 0.05 and a |log2(fold-change)| > 1 as significance thresholds [15] [38].
Candidate lncRNA Selection via Survival Analysis
- Univariate Analysis: Perform univariate Cox regression on the DElncs using disease-free survival (DFS) or overall survival (OS) as the endpoint. Retain lncRNAs with a significance level of P < 0.05 [43] [15]. This yields a refined pool of recurrence-related dysregulated lncRNAs for subsequent analysis.
Application of Machine Learning for Feature Selection
This step involves applying multiple feature selection methods to the candidate lncRNAs to identify a robust subset.
- LASSO Cox Regression: Execute LASSO regression using the R package
glmnet. Perform 10-fold cross-validation to determine the optimal value of the penalty parameter (lambda) that minimizes the cross-validation error. The lncRNAs with non-zero coefficients at this lambda are selected [43] [15] [44]. - Random Forest: Run the Random Forest algorithm using the R package
randomForest. Rank all candidate lncRNAs by their importance value (mean decrease in accuracy or Gini). Select the top-ranked features (e.g., top 30) for further consideration [15]. - SVM-RFE: Implement SVM-RFE using the R package
e1071. Utilize a linear kernel and 5-fold cross-validation. The algorithm will recursively eliminate features and output an optimal feature subset based on predictive accuracy [15] [38]. - Integration of Results: Identify the final candidate lncRNAs by taking the intersection of the features selected by at least two of the three machine learning methods. A Venn diagram is a useful tool for this step [15].
Multivariate Model Building and Validation
- Signature Construction: Perform multivariate Cox proportional hazards regression on the final candidate lncRNAs. Use the resulting coefficients to calculate a risk score for each patient:
Risk Score = Σ (lncRNA_coefficient_i × lncRNA_expression_i)[43] [15]. - Performance Evaluation:
- ROC Analysis: Assess the signature's predictive power for recurrence at specific time points (e.g., 1, 2, 3 years) using time-dependent Receiver Operating Characteristic (ROC) analysis in the training cohort [43] [15].
- Survival Analysis: Divide patients into high-risk and low-risk groups based on the median risk score from the training set. Use Kaplan-Meier survival analysis and the log-rank test to compare disease-free survival between the two groups in both the training and independent validation cohorts [43] [15].
- Independent Validation: Confirm the prognostic performance of the signature in the held-out validation cohort and, if available, in an external patient cohort [15].
Diagram 1: Integrated workflow for lncRNA signature development using multiple machine learning feature selection techniques.
Successful execution of the described protocols relies on a suite of specific computational tools, data resources, and experimental reagents.
Table 2: Key Research Reagent Solutions for lncRNA Biomarker Discovery
| Category | Item | Specific Example / Catalog Number | Critical Function in Workflow |
|---|---|---|---|
| Data Resources | TCGA-LIHC Database | https://portal.gdc.cancer.gov/ | Primary source of lncRNA expression and clinical data for model training [43] [15] [38] |
| Software & Packages | R Statistical Software | v3.3.3 or higher | Core platform for data analysis, statistics, and model building [15] [38] |
| Bioinformatic R Packages | glmnet, randomForest, e1071, survival, DESeq2, edgeR, limma |
Implementation of specific algorithms for differential expression, feature selection, and survival analysis [43] [15] [38] | |
| Wet-Lab Reagents | RNA Extraction Kit | miRNeasy Mini Kit (QIAGEN, 217004) / Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit (Norgen Biotek) [6] [25] | Isolates high-quality total RNA from tissues or liquid biopsy samples (plasma) |
| cDNA Synthesis Kit | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622) [6] | Generates complementary DNA from purified RNA for downstream qPCR | |
| qRT-PCR Master Mix | Power SYBR Green PCR Master Mix (Thermo Fisher) [6] [25] | Enables quantitative measurement of lncRNA expression levels | |
| Reference Genes | Endogenous Control | GAPDH, β-actin, SNORD72, U6 [14] [6] [25] | Normalizes lncRNA expression data to account for technical variability |
Concluding Remarks
The strategic integration of LASSO, Random Forest, and SVM-RFE provides a powerful, multi-faceted approach for pinpointing critical lncRNAs from high-dimensional datasets. LASSO delivers sparse models ideal for clinical translation, Random Forest robustly handles complex biological interactions, and SVM-RFE excels at defining optimal diagnostic feature sets. Following the detailed protocols and utilizing the referenced toolkit will equip researchers to develop validated, clinically relevant lncRNA signatures, thereby advancing the integration of machine learning into molecular diagnostics for HCC and solidifying the foundation for personalized medicine in oncology.
The integration of machine learning (ML) into Hepatocellular Carcinoma (HCC) research represents a paradigm shift from conventional diagnostic approaches, enabling the analysis of complex molecular signatures like long non-coding RNA (lncRNA) biomarkers alongside clinical data. The development of HCC is an intricate process involving liver injury, chronic inflammation, fibrosis, and cirrhosis, with various molecular impairments like microRNA dysregulation and immunomodulation contributing to its pathogenesis [14]. Current diagnostic standards, which rely on serum alpha-fetoprotein (AFP) levels and imaging techniques, demonstrate limited sensitivity and specificity, particularly for early-stage detection [14]. Machine learning algorithms address these limitations by identifying multidimensional patterns in heterogeneous data sources, facilitating earlier and more accurate diagnosis. This document provides a comprehensive overview of four key ML algorithms—LightGBM (LGBM), Support Vector Machines (SVM), Random Forest (RF), and Neural Networks (NN)—within the context of constructing robust diagnostic models for HCC, with particular emphasis on their application to lncRNA biomarker integration.
Core Algorithm Characteristics
The selection of an appropriate machine learning algorithm is critical for developing effective HCC diagnostic models. Each algorithm possesses distinct mechanistic strengths that determine its suitability for processing complex biomarker data.
LightGBM (LGBM): A gradient boosting framework that excels in speed and efficiency through histogram-based algorithms and two innovative techniques: Gradient-based One-Side Sampling (GOSS) and Exclusive Feature Bundling (EFB) [45] [46]. GOSS prioritizes data instances with larger gradients during training, thereby focusing computational resources on difficult-to-predict cases and improving training efficiency without significantly distorting the data distribution [46]. EFB identifies mutually exclusive features (those rarely taking non-zero values simultaneously) and bundles them into a single feature, effectively reducing dimensionality and accelerating model training [45]. This architecture is particularly advantageous for high-dimensional genomic data, making it ideal for integrating numerous lncRNA biomarkers with standard clinical parameters.
Support Vector Machines (SVM): This algorithm operates on the principle of identifying an optimal hyperplane that maximizes the margin between different classes in the data [47]. For non-linearly separable data, SVM employs the kernel trick, which implicitly maps input features into higher-dimensional spaces where effective linear separation becomes possible [48] [47]. While effective in high-dimensional spaces, its performance is highly sensitive to parameter selection (e.g., regularization parameter C and kernel parameters), and it can become computationally intensive with large datasets [48].
Random Forest (RF): An ensemble method that constructs multiple decision trees during training and outputs the mode of their classes (for classification) or mean prediction (for regression) [49]. Its robustness stems from feature bagging—where each tree is built using a random subset of features—and aggregation of predictions from all trees [49] [50]. This approach reduces overfitting risk, a common issue with single decision trees, and provides native feature importance estimation [50]. RF can handle datasets with missing values effectively, making it suitable for real-world clinical data that often contains incomplete records [49] [50].
Neural Networks (NN): These are complex networks of interconnected artificial neurons that learn hierarchical representations of data through successive layers of processing [51] [52]. Their multi-layered structure (input, hidden, and output layers) enables modeling of highly non-linear relationships through forward propagation of data and backpropagation of errors to adjust internal weights [52]. This architectural flexibility makes them particularly powerful for identifying intricate patterns across diverse data types, from clinical parameters to complex lncRNA expression profiles.
Quantitative Performance in HCC Detection
Recent clinical studies demonstrate the substantial potential of these algorithms, particularly LGBM and RF, in HCC detection workflows. The following table summarizes key performance metrics from recent clinical validation studies:
Table 1: Comparative Performance of ML Algorithms in HCC Detection
| Algorithm | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | Study Cohort |
|---|---|---|---|---|---|
| LGBM | 98.75 [14] | 94.9 [53] | 99.5 [53] | 0.99 [53] | Filipino [53] & Egyptian [14] |
| Random Forest | 98.9 [53] | 90.5 [53] | 99.8 [53] | 0.99 [53] | Filipino [53] |
| Neural Networks | 91.25 [14] | Not Reported | Not Reported | Not Reported | Egyptian [14] |
| SVM | 88.75 [14] | Not Reported | Not Reported | Not Reported | Egyptian [14] |
| k-NN | 87.50 [14] | Not Reported | Not Reported | Not Reported | Egyptian [14] |
These results highlight the superior performance of tree-based ensemble methods (LGBM and RF) in HCC detection tasks. Notably, a study on a Filipino cohort achieved high predictive performance using only seven clinical predictors: age, albumin, alkaline phosphatase (ALP), alpha-fetoprotein (AFP), des-gamma-carboxy prothrombin (DCP), aspartate transaminase, and platelet count [53]. This streamlined predictor set is particularly advantageous for resource-limited settings, demonstrating how ML can optimize diagnostic efficiency.
Experimental Protocols for HCC Model Development
Workflow for ML-Based HCC Detection
A standardized workflow ensures reproducible development of HCC diagnostic models, from initial data collection through final model validation. The following diagram illustrates the comprehensive protocol for constructing and validating ML models for HCC detection:
Diagram 1: Comprehensive workflow for ML-based HCC detection model development
Data Collection & Preprocessing Protocol
Patient Cohort Selection: In a recent study, researchers enrolled 267 subjects classified into 98 healthy controls, 67 with benign liver conditions, and 102 with HCC [14]. All participants provided written informed consent, and the study was approved by the institutional ethical committee following REMARK guidelines [14].
Clinical & Molecular Data Acquisition: Collect comprehensive clinico-demographic data (age, sex, smoking history, cirrhosis status) and serum parameters (ALT, AST, bilirubin, albumin, INR, AFP, HBV/HCV antibodies) [14]. For lncRNA analysis, purify total RNA from serum samples using a miRNEasy extraction kit (Qiagen) [14]. Validate RNA quality and purity using a Qubit 3.0 Fluorimeter with appropriate assay kits [14].
Feature Selection: Apply multiple feature selection techniques (Pearson correlation, random forest feature selection, information gain, recursive feature elimination, Lasso regression) to identify the most predictive variables [53]. Studies have demonstrated that only 7-10 key predictors may be sufficient for high-accuracy detection, including age, albumin, ALP, AFP, DCP, AST, and platelet count [53].
Model Training & Validation Protocol
Algorithm Implementation: Implement multiple algorithms (KNN, RF, SVM, LGBM, DNNs) using standard ML libraries (e.g., scikit-learn for Python). For LGBM, initialize with
LGBMClassifier(learning_rate=0.09, max_depth=-5, random_state=42)and fit with evaluation metrics and validation sets to monitor training [45].Hyperparameter Optimization: Determine optimal hyperparameters using a grid-search approach with cross-validation [53]. For LGBM, key parameters include
boosting_type('gbdt', 'dart', or 'goss'),num_leaves,learning_rate,max_depth, and regularization parameters (lambda_l1,lambda_l2) [46].Performance Validation: Evaluate models using standard metrics: accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and area under the receiver operating characteristic curve (AUC) [53]. Employ k-fold cross-validation and hold-out test sets to ensure robustness and generalizability.
LncRNA Biomarkers in HCC Pathogenesis
The integration of lncRNA biomarkers with machine learning represents a cutting-edge approach for HCC diagnosis. Research has identified several key lncRNAs involved in HCC pathogenesis, particularly through their interactions with autophagy and cytokine signaling pathways. The following diagram illustrates the molecular relationships between these biomarkers:
Diagram 2: Molecular interactions of lncRNA biomarkers in HCC pathogenesis
The pathway illustrates how differentially expressed lncRNAs (lncRNA-RP11-513I15.6 and lncRNA-WRAP53) interact with microRNAs (miR-1262, miR-1298, and miR-106b-3p) to regulate key mRNAs (RAB11A, STAT1, and ATG12) involved in autophagy and cytokine signaling processes central to HCC development [14]. These molecular interactions form a complex regulatory network that machine learning models can exploit for highly specific HCC detection.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for HCC Biomarker Studies
| Reagent / Kit | Manufacturer | Function in HCC Research |
|---|---|---|
| miRNEasy Extraction Kit | Qiagen | Purification of total RNA (including small RNAs) from serum or tissue samples [14] |
| Qubit TM RNA HS Assay Kit | Invitrogen | Validation of RNA quality, purity, and concentration using fluorometric quantification [14] |
| miScript II RT Kit | Qiagen | Reverse transcription of purified RNA for subsequent qRT-PCR analysis [14] |
| Quantitect SYBR Green Master Mix | Qiagen | qRT-PCR quantification of mRNA expression levels (e.g., RAB11A, STAT1, ATG12) [14] |
| miScript SYBR Green PCR Kit | Qiagen | qRT-PCR quantification of miRNA expression levels (e.g., miR-1262, miR-1298) [14] |
| RT2 SYBR Green ROX qPCR Master mix | Qiagen | qRT-PCR quantification of lncRNA expression levels (e.g., lncRNA-RP11-513I15.6) [14] |
| Chrysosplenetin | Chrysosplenetin|Natural O-Methylated Flavonol for Research | High-purity Chrysosplenetin for research. Explore its applications in osteogenesis, cancer, and anti-malarial studies. This product is For Research Use Only. Not for human use. |
| Moxifloxacin hydrochloride monohydrate | Moxifloxacin hydrochloride monohydrate, CAS:192927-63-2, MF:C21H27ClFN3O5, MW:455.9 g/mol | Chemical Reagent |
The integration of machine learning with lncRNA biomarker analysis represents a transformative approach for HCC diagnosis, offering significant improvements over conventional diagnostic methods. Among the algorithms evaluated, LightGBM and Random Forest consistently demonstrate superior performance in clinical validation studies, achieving accuracy rates exceeding 98% in diverse patient populations [53] [14]. Their efficiency in handling high-dimensional data, native support for feature importance analysis, and robustness against overfitting make them particularly suitable for integrating complex molecular signatures with standard clinical parameters. The experimental protocols and reagent solutions outlined provide a reproducible framework for researchers developing HCC diagnostic models. As the field advances, the synergy between molecular biomarker discovery and optimized machine learning algorithms will undoubtedly enhance early detection capabilities, ultimately improving patient outcomes in hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) represents a significant global health challenge, ranking as the sixth most prevalent cancer and the third leading cause of cancer-related mortality worldwide [37] [54]. The insidious nature of HCC progression, coupled with limited early diagnostic tools, results in a majority of patients being diagnosed at advanced stages when curative treatment options are no longer viable [54]. Despite being the current golden standard for HCC screening, alpha-fetoprotein (AFP) testing demonstrates limited sensitivity and specificity, highlighting the urgent need for more reliable biomarkers [55] [56] [6].
Long non-coding RNAs (lncRNAs) have emerged as promising molecular biomarkers in oncology. These transcripts, exceeding 200 nucleotides in length without protein-coding capacity, are intensively involved in HCC progression through diverse mechanisms including epigenetic regulation, microRNA sponging, and modulation of key signaling pathways [37] [56]. The stability of lncRNAs in bodily fluids, combined with their cancer-specific expression patterns, positions them as ideal candidates for minimally invasive liquid biopsy approaches [25] [54].
The integration of machine learning algorithms into biomarker discovery has revolutionized the identification and validation of lncRNA signatures. This computational approach enables analysis of high-dimensional transcriptomic data to identify optimal biomarker combinations with enhanced predictive power [37] [55]. This application note examines successful case studies implementing lncRNA-based biomarkers for HCC, detailing experimental protocols and analytical frameworks to guide researchers in this rapidly advancing field.
Case Studies: lncRNA Signatures in HCC
Case Study 1: A 4-lncRNA Signature for Predicting Early Recurrence
Background and Rationale: Nearly 70% of HCC patients experience postoperative recurrence within five years, with most cases representing early recurrence (within two years of surgery) associated with significantly reduced five-year survival rates [37]. Predicting this early recurrence would enable improved surveillance strategies and personalized adjuvant therapy approaches.
Signature Identification and Performance: Researchers analyzed RNA expression data from 314 HCC patients with complete survival records from the TCGA-LIHC database. Through a rigorous analytical pipeline combining three differential expression methods (DESeq2, edgeR, and limma) and two survival analyses (log-rank and Cox methods), they identified 81 recurrence-associated differentially expressed lncRNAs [37].
Machine learning refinement employing three algorithms - Least Absolute Shrinkage and Selection Operator (LASSO), Random Forest, and Support Vector Machine Recursive Feature Elimination (SVM-RFE) - narrowed candidates to 11 lncRNAs. Subsequent multivariate Cox analysis yielded a final signature of four lncRNAs: AC108463.1, AF131217.1, CMB9-22P13.1, and TMCC1-AS1 [37].
Table 1: The 4-lncRNA Signature for HCC Early Recurrence Prediction
| lncRNA | Expression in HCC | Risk Association | Functional Role |
|---|---|---|---|
| AC108463.1 | Not specified | High-risk | Mechanism not fully elucidated |
| AF131217.1 | Not specified | High-risk | Mechanism not fully elucidated |
| CMB9-22P13.1 | Not specified | High-risk | Mechanism not fully elucidated |
| TMCC1-AS1 | Not specified | High-risk | Mechanism not fully elucidated |
The risk score was calculated using the formula:
Risk Score = (0.1916 × AC108463.1) + (2.2304 × AF131217.1) + (0.3156 × CMB9-22P13.1) + (0.2476 × TMCC1-AS1)
Patients stratified into high-risk and low-risk groups based on the median risk score showed significantly different early recurrence rates, with the high-risk group demonstrating markedly poorer outcomes. The signature's predictive performance was further enhanced when combined with established clinical markers (AFP, TNM stage), and validation in an external cohort of 44 patients from Jinling Hospital confirmed its clinical utility [37].
Biological Insights: Gene set enrichment analysis revealed several molecular pathways associated with HCC pathogenesis were enriched in the high-risk group. Additionally, antitumor immune cells (activated B cells, type 1 T helper cells, natural killer cells, and effective memory CD8 T cells) were enriched in the low-risk group, suggesting distinct immune microenvironments between the subgroups [37].
Case Study 2: A 9-lncRNA Diagnostic Panel for HBV-Related HCC
Background and Rationale: Hepatitis B virus (HBV) infection represents a major risk factor for HCC development, accounting for a substantial proportion of cases worldwide. The distinct molecular pathogenesis of HBV-related HCC warrants the development of etiology-specific diagnostic biomarkers.
Signature Identification and Performance: This study implemented a comprehensive bioinformatics approach to identify lncRNA biomarkers specific for HBV-related HCC. Researchers analyzed expression profiles from three GEO datasets (GSE55092, GSE19665, and GSE84402), identifying 38 differentially expressed lncRNAs and 543 differentially expressed mRNAs in HBV-related HCC tissues compared to non-tumor controls [57].
Machine learning feature selection identified nine optimal diagnostic lncRNA biomarkers: AL356056.2, AL445524.1, TRIM52-AS1, AC093642.1, EHMT2-AS1, AC003991.1, AC008040.1, LINC00844, and LINC01018. The support vector machine (SVM) model achieved an area under the curve (AUC) of 0.957 with 95.7% specificity and 100% sensitivity, while the random forest model achieved an AUC of 0.904 with 94.3% specificity and 86.5% sensitivity [57].
Table 2: The 9-lncRNA Diagnostic Panel for HBV-Related HCC
| lncRNA | Expression Pattern | Diagnostic Performance | Clinical Utility |
|---|---|---|---|
| AL356056.2 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| AL445524.1 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| TRIM52-AS1 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| AC093642.1 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| EHMT2-AS1 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| AC003991.1 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| AC008040.1 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| LINC00844 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
| LINC01018 | Not specified | Contributed to SVM model (AUC=0.957) | HBV-related HCC diagnosis |
Functional Implications: Co-expression network analysis and functional annotation revealed that the target differentially expressed mRNAs were enriched in key carcinogenic pathways including the p53 signaling pathway, retinol metabolism, PI3K-Akt signaling cascade, and chemical carcinogenesis. This suggests these lncRNAs may modulate inflammatory conditions in the tumor immune microenvironment of HBV-related HCC [57].
Additional Notable lncRNA Signatures in HCC
Several other studies have developed lncRNA-based signatures with prognostic and diagnostic value in HCC:
- A costimulatory molecule-related 5-lncRNA signature (BOK-AS1, AC099850.3, AL365203.2, NRAV, and AL049840.4) demonstrated significant prognostic power, with high-risk patients showing shorter overall survival times [58].
- An autophagy-related 4-lncRNA signature (LUCAT1, AC099850.3, ZFPM2-AS1, and AC009005.1) served as an independent prognostic indicator for HCC patients, with AUC values of 0.764, 0.738, and 0.717 for 1-, 3-, and 5-year survival, respectively [59].
- A plasma-based detection of four lncRNAs (LINC00152, LINC00853, UCA1, and GAS5) integrated with conventional laboratory parameters through machine learning achieved 100% sensitivity and 97% specificity for HCC diagnosis [6].
- For advanced chronic hepatitis C patients, plasma lncRNAs HULC and RP11-731F5.2 were identified as potential biomarkers for HCC risk assessment [25].
Experimental Protocols
Sample Collection and RNA Extraction
Patient Selection and Ethical Considerations:
- Obtain written informed consent from all participants following protocol approval by institutional ethics committees [6] [25].
- For HCC patients, confirm diagnosis through LI-RADS imaging criteria or histopathological examination of tissue biopsies [6].
- Include appropriate control groups (healthy individuals, patients with benign liver conditions, or paracancerous tissues) matched for age and gender [6] [25].
- Collect clinical data including etiology, liver function tests, AFP levels, imaging characteristics, and pathological staging [6].
Sample Collection and Processing:
- Collect peripheral blood in EDTA-containing tubes and process within 2 hours of collection [25].
- Centrifuge blood samples at 704 × g for 10 minutes to separate plasma [25].
- Aliquot plasma samples and store at -70°C until RNA extraction to prevent degradation [25].
- For tissue samples, snap-freeze in liquid nitrogen immediately following surgical resection and store at -80°C [59].
RNA Extraction:
- Extract total RNA from 500 μL plasma using specialized kits for circulating RNA (e.g., Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit) [25].
- For tissue samples or cell lines, use TRIzol reagent according to manufacturer protocols [59].
- Treat RNA samples with DNase to remove genomic DNA contamination [25].
- Quantify RNA quality and concentration using spectrophotometry or bioanalyzer systems.
cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR)
Reverse Transcription:
- Use High-Capacity cDNA Reverse Transcription Kit with 500 ng-1 μg total RNA as template [25].
- Include controls without reverse transcriptase to assess genomic DNA contamination.
- Perform reactions according to manufacturer protocols using a thermal cycler.
qRT-PCR Analysis:
- Use Power SYBR Green PCR Master Mix according to manufacturer protocols [6] [25].
- Design primers specifically targeting lncRNAs of interest (see Table 3 for examples).
- Perform reactions in triplicate on a real-time PCR system (e.g., StepOne Plus System or ViiA 7 system) [6] [25].
- Use the following cycling conditions: initial denaturation at 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 60-62°C for 1 minute [25].
- Include no-template controls in each run to monitor for contamination.
- Normalize expression data using reference genes (e.g., GAPDH or β-actin) and calculate relative expression using the 2−ΔΔCt method [6] [25].
Table 3: Example Primer Sequences for lncRNA Detection
| lncRNA | Forward Primer (5'-3') | Reverse Primer (5'-3') | Reference |
|---|---|---|---|
| AC099850.3 | TCGCTATGTTTCCCAGGCTG TATT | TGCCAAGGAATCTCTGAAGT CCAT | [59] |
| LUCAT1 | GTGTCCAAATGCTGTCCCTCA TCTC | ATCCTCGGGTTGCCTCTGTT TA | [59] |
| ZFPM2-AS1 | TGGTGGTATTTCTGCTGTTC TC | GTTCCATCTTCCTCCTTGTC TAC | [59] |
| GAPDH | ACCCACTCCTCCACCTTTGAC | TGTTGCTGTAGCCAAATTCG TT | [59] |
Bioinformatics and Machine Learning Analysis
Data Acquisition and Preprocessing:
- Download RNA expression data from public databases (TCGA, GEO, exoRBase) [37] [55] [60].
- Normalize data using appropriate methods (e.g., TPM for RNA-seq data) [55].
- Remove features not expressed in more than 80% of samples to reduce noise [55].
- Scale data by sample to unit l2-norm to maximize accuracy and reduce fit time [55].
Differential Expression Analysis:
- Identify differentially expressed lncRNAs using R packages such as "DESeq2", "edgeR", or "limma" [37].
- Apply filtering criteria (e.g., |log2FC| > 1-2 and FDR < 0.05) to identify significant differentially expressed lncRNAs [37] [60].
Feature Selection and Model Construction:
- Apply univariate Cox regression to identify lncRNAs associated with survival outcomes [58] [60].
- Use machine learning algorithms (LASSO, SVM-RFE, Random Forest) for dimensionality reduction and feature selection [37] [55].
- Perform multivariate Cox regression to finalize signature lncRNAs and calculate coefficients [37] [58].
- Split data into training, validation, and test sets (typically 70-80% for training) in a stratified manner [55].
Model Validation:
- Perform internal validation using bootstrap resampling or cross-validation [59].
- Validate signatures in external independent cohorts when possible [37].
- Evaluate performance using time-dependent ROC curves, Kaplan-Meier survival analysis, and concordance indices [37] [58].
- Compare with existing clinical biomarkers and staging systems to assess added value.
Visualizing Experimental Workflows and Signaling Pathways
Workflow for lncRNA Signature Development
Machine Learning Approach for Biomarker Discovery
The Scientist's Toolkit: Essential Research Reagents
Table 4: Essential Research Reagents for lncRNA Biomarker Studies
| Reagent/Kits | Specific Example | Application Purpose | Key Considerations |
|---|---|---|---|
| RNA Extraction Kit | Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit (Norgen Biotek) | Isolation of high-quality RNA from plasma samples | Optimized for low-abundance circulating RNA |
| Reverse Transcription Kit | High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher) | cDNA synthesis from RNA templates | Includes RNase inhibitor for improved yield |
| qPCR Master Mix | Power SYBR Green PCR Master Mix (Thermo Fisher) | Quantitative detection of lncRNAs | Provides consistent amplification efficiency |
| Cell Culture Media | DMEM with 10% FBS and antibiotics | Maintenance of HCC cell lines | Ensure optimal growth conditions for experiments |
| Bioinformatics Tools | R packages: "edgeR", "DESeq2", "limma", "glmnet", "randomForest" | Differential expression and machine learning analysis | Use latest versions for updated algorithms |
| Clinical Data Management | SPSS, GraphPad Prism | Statistical analysis and visualization | Facilitates correlation with clinical parameters |
| Econazole Nitrate | Econazole Nitrate | High-purity Econazole Nitrate for life science research. A broad-spectrum synthetic antifungal compound. For Research Use Only. Not for human or veterinary use. | Bench Chemicals |
| Sideroxylin | Sideroxylin | C18H16O5 | CAS 3122-87-0 | Bench Chemicals |
The integration of lncRNA biomarkers and machine learning algorithms represents a transformative approach in HCC diagnostics and prognostics. The case studies presented demonstrate that multi-lncRNA signatures consistently outperform single biomarkers in predicting clinical outcomes, with machine learning playing a pivotal role in identifying optimal biomarker combinations from high-dimensional data.
Future developments in this field will likely focus on validating these signatures in large, multi-center prospective cohorts and standardizing detection protocols for clinical implementation. Additionally, incorporating lncRNA signatures into composite models that include protein biomarkers, clinical parameters, and imaging characteristics will further enhance their clinical utility. As our understanding of lncRNA biology expands, these molecular signatures promise to significantly improve early detection, prognostic stratification, and personalized treatment approaches for hepatocellular carcinoma.
Hepatocellular carcinoma (HCC) remains a significant global health challenge, characterized by late-stage diagnosis and poor prognosis. The integration of long non-coding RNA (lncRNA) expression profiles with established clinical data—including alpha-fetoprotein (AFP) levels, TNM staging, and liver function tests—represents a transformative approach for enhancing diagnostic precision and prognostic assessment in HCC management. This protocol outlines standardized methodologies for generating and integrating multi-dimensional data to construct robust predictive models, advancing the broader thesis of machine learning-enabled lncRNA biomarker integration for HCC diagnosis.
Quantitative Data Synthesis for Integrated Analysis
Table 1: Performance Metrics of Individual lncRNAs and Integrated Models in HCC Diagnosis
| Biomarker / Model | Sensitivity (%) | Specificity (%) | AUC | Clinical Correlation | Reference |
|---|---|---|---|---|---|
| LINC00152 | 83 | 67 | 0.79 | Positive correlation with tumor proliferation [6] | [6] |
| GAS5 | 60 | 53 | 0.62 | Inverse correlation with mortality risk [6] | [6] |
| LINC00853 | 77 | 60 | 0.72 | Associated with HCC progression [6] | [6] |
| UCA1 | 73 | 57 | 0.68 | Promotes cell proliferation and inhibits apoptosis [6] | [6] |
| LINC00152/GAS5 Ratio | N/A | N/A | N/A | Significant correlation with increased mortality risk [6] | [6] |
| 10-core EV-derived lncRNA Panel | N/A | N/A | N/A | Association with HCC progression via autophagy/MAPK pathways [61] | [61] |
| Machine Learning Model | 100 | 97 | ~1.00 | Superior to individual biomarkers [6] | [6] |
Table 2: Correlation of AFP Status with HCC Clinicopathological Features
| Clinical Parameter | AFP-Negative (<20 ng/mL) | AFP-Positive (≥20 ng/mL) | P-value |
|---|---|---|---|
| Well/Moderately Differentiated Tumors | 34.0% | 66.0% | <0.001 [62] |
| Poorly Differentiated/Anaplastic Tumors | 17.0% | 83.0% | <0.001 [62] |
| TNM Stage I/II | 36.2% | 63.8% | <0.001 [62] |
| Tumor Size ≤5 cm | 36.3% | 63.7% | <0.001 [62] |
| 5-Year Survival (No Surgery) | Better | Poorer | <0.001 [62] |
Experimental Protocols
Protocol for Serum/Plasma Collection and EV-Derived lncRNA Analysis
Principle: Extracellular vesicles (EVs) contain disease-specific RNA signatures that offer promising avenues for non-invasive biomarker discovery [61].
Reagents and Equipment:
- Vacuum tubes with inert separation gel and procoagulant (for serum)
- EDTA anticoagulant tubes (for plasma)
- 0.8 μm filters
- Gel-permeation column (ES911, Echo Biotech)
- 100kD ultrafiltration tubes
- RNA Purification Kit (Simgen, cat. 5202050)
Procedure:
- Sample Collection: Collect fasting venous blood from patients and controls prior to treatment initiation.
- Processing: Centrifuge samples within 2 hours of collection. Separate serum/plasma and aliquot into sterile tubes.
- Storage: Store aliquots at -80°C until EV isolation.
- EV Isolation: a. Thaw samples and pretreat with 0.8 μm filter b. Separate via gel-permeation column c. Collect PBS eluent from tubes 7-9 d. Concentrate using 100kD ultrafiltration tube
- EV Characterization: a. Analyze particle size distribution by nano-flow cytometry b. Examine morphology by transmission electron microscopy with uranyl acetate staining c. Confirm marker proteins (TSG101, Alix, CD9) and negative control (Calnexin) by Western blot [61]
- RNA Extraction: a. Add 700 µL Buffer TL and 100 µL Buffer EX to 100 µL EV suspension b. Vortex and centrifuge (12,000 × g, 4°C, 15 min) c. Combine supernatant with ethanol and load onto purification column d. Centrifuge (12,000 × g, 30 s), discard flow-through e. Wash column with Buffer WA and Buffer WBR (12,000 × g, 30 s each) f. Air-dry column (14,000 × g, 1 min) g. Elute RNA with 35 µL RNase-free water [61]
Protocol for Plasma lncRNA Quantification via qRT-PCR
Principle: Circulating lncRNAs in plasma serve as accessible biomarkers for liquid biopsy in HCC [6].
Reagents and Equipment:
- miRNeasy Mini Kit (QIAGEN, cat no. 217004)
- RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, cat no. K1622)
- PowerTrack SYBR Green Master Mix (Applied Biosystems, cat no. A46012)
- ViiA 7 real-time PCR system (Applied Biosystems)
- Primers for target lncRNAs (LINC00152, LINC00853, UCA1, GAS5) and housekeeping gene (GAPDH)
Procedure:
- RNA Isolation: Extract total RNA from plasma samples using miRNeasy Mini Kit according to manufacturer's protocol.
- cDNA Synthesis: Perform reverse transcription using RevertAid First Strand cDNA Synthesis Kit on a thermal cycler.
- qRT-PCR Setup: a. Prepare reactions using PowerTrack SYBR Green Master Mix b. Set up reactions in triplicate for each sample c. Run on ViiA 7 real-time PCR system with appropriate cycling conditions
- Data Analysis: a. Calculate relative quantification using the ΔΔCT method b. Normalize to GAPDH expression c. Determine expression ratios (e.g., LINC00152 to GAS5 ratio) [6]
Protocol for Integrated Data Analysis Using Machine Learning
Principle: Machine learning algorithms can effectively integrate lncRNA expression with clinical parameters to improve HCC diagnosis and prognosis [9] [6].
Software and Tools:
- Python Scikit-learn platform
- lncRNACNVIntegrateR package for R [63]
- Statistical software (e.g., Minitab, R)
Procedure:
- Data Compilation: a. Create structured dataset with lncRNA expression values (LINC00152, LINC00853, UCA1, GAS5) b. Incorporate clinical parameters: AFP levels, TNM stage, liver function tests (ALT, AST, bilirubin, albumin), tumor size, demographic data
- Feature Engineering: a. Calculate lncRNA ratios (e.g., LINC00152/GAS5) b. Normalize continuous variables c. Encode categorical variables (TNM stage, etc.)
- Model Training: a. Implement multiple algorithms (Random Forest, XGBoost, SVM, neural networks) b. Utilize training-validation split (e.g., 70:30) c. Optimize hyperparameters via cross-validation
- Model Validation: a. Assess performance using ROC analysis, sensitivity, specificity b. Validate in independent cohort when available c. Perform decision curve analysis to evaluate clinical utility [6] [64]
Visual Integration Workflows
Integrated Data Analysis Workflow
lncRNA-Clinical Parameter Regulatory Network
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Kits for Integrated lncRNA-Clinical Studies
| Reagent/Kits | Function | Application Example | Key Features |
|---|---|---|---|
| miRNeasy Mini Kit (QIAGEN) | Total RNA isolation from plasma/serum | Plasma lncRNA extraction for qRT-PCR | Maintains RNA integrity; includes DNase treatment [6] |
| RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) | Reverse transcription for cDNA synthesis | Preparation of templates for lncRNA quantification | High efficiency with complex RNA samples [6] |
| PowerTrack SYBR Green Master Mix (Applied Biosystems) | qRT-PCR detection | lncRNA expression quantification | Optimized for difficult targets; high sensitivity [6] |
| RNA Purification Kit (Simgen) | EV-RNA extraction | Isolation of RNA from extracellular vesicles | Specifically designed for EV RNA recovery [61] |
| Size-Exclusion Chromatography Columns (Echo Biotech) | EV isolation and purification | Separation of EVs from biofluids | Preserves EV integrity and biomolecule content [61] |
| lncRNACNVIntegrateR Package | Multi-omics data integration | Correlating lncRNA expression with CNV and clinical data | User-friendly R package for integrative analysis [63] |
| Cochlioquinone A | Cochlioquinone A | Natural Product for Research | Cochlioquinone A is a fungal metabolite & zinc ionophore for autophagy, immunology, and antifungal research. For Research Use Only. | Bench Chemicals |
Data Interpretation Guidelines
Expression Patterns: Elevated oncogenic lncRNAs (LINC00152, UCA1) with suppressed tumor-suppressive lncRNAs (GAS5) typically indicate aggressive HCC phenotypes [6].
AFP Integration: In AFP-negative cases, lncRNA signatures provide critical diagnostic information; combinations significantly improve detection sensitivity [6] [64].
Staging Correlation: lncRNA expression profiles often correlate with TNM stage - more advanced stages typically show more dysregulated lncRNA patterns [61] [62].
Prognostic Assessment: Ratios such as LINC00152/GAS5 provide superior prognostic information compared to individual markers alone [6].
Therapeutic Implications: Identified lncRNA signatures can inform therapeutic targets, as many lncRNAs regulate key pathways in HCC progression (e.g., MAPK, autophagy) [61] [11].
This comprehensive protocol provides researchers with standardized methodologies for integrating lncRNA biomarkers with conventional clinical data, facilitating the development of more accurate diagnostic and prognostic models for hepatocellular carcinoma.
Navigating Challenges: Data Biases, Model Interpretability, and Clinical Translation Roadblocks
The integration of machine learning (ML) with long non-coding RNA (lncRNA) biomarkers represents a transformative frontier in hepatocellular carcinoma (HCC) diagnostics. However, the development of robust, clinically applicable models faces significant methodological challenges rooted in dataset limitations. Issues such as biased training cohorts, inadequate sample sizes, and failure to account for competing clinical risks fundamentally compromise model validity and generalizability [65]. This application note provides a structured framework to overcome these limitations, enabling the development of HCC diagnostic models that maintain predictive accuracy across diverse clinical populations. We present standardized protocols for bias mitigation, data augmentation, and model validation specifically tailored to lncRNA biomarker research, providing researchers with practical tools to enhance the reliability of their predictive models.
Quantitative Landscape of Current HCC Prediction Approaches
Table 1: Performance Metrics of Selected HCC Prediction Models
| Model Type | Key Features/Factors | Sample Size | Performance | Reference/Context |
|---|---|---|---|---|
| EV-derived lncRNA Signature | 10 core lncRNAs; lncRNA-miRNA-mRNA network | 24 participants (discovery) | Identified 133 significantly differentially expressed lncRNAs | [61] |
| Machine Learning (with feature reduction) | Feature reduction via RFE, PCA | Not specified | Accuracy: 94.67%-97.33% (various algorithms) | [66] |
| ML for MASLD-HCC Risk | FIB-4 score as key predictor | 1,561 (training), 686 (validation) | AUC: 0.97, Accuracy: 92.06%, Sensitivity: 74.41% | [67] |
| AI-Ultrasound Screening | UniMatch (detection) & LivNet (classification) | 17,913 images (training) | Sensitivity: 0.956, Specificity: 0.787 (Strategy 4) | [68] |
| GALAD Serum Biomarker | Gender, Age, AFP-L3, AFP, DCP | 1,558 patients with cirrhosis | AUC: 0.78 (vs. 0.66 for AFP alone) | [69] |
| Competing Risk Analysis | Fine-Gray vs. Cox regression | 1,629 patients | Mean 3-year HCC risk: 3.24% (Fine-Gray) vs. 3.37% (Cox) | [65] |
Table 2: Impact of Feature Reduction on Machine Learning Performance for HCC Prediction
| Machine Learning Algorithm | Accuracy Before Feature Reduction | Accuracy After Feature Reduction |
|---|---|---|
| Naive Bayes | Not specified | 97.33% |
| Support Vector Machine (SVM) | Not specified | 96.00% |
| Neural Networks | Not specified | 96.00% |
| Decision Tree | Not specified | 96.00% |
| K-Nearest Neighbors (KNN) | 70.6% (on original dataset) | 94.67% |
Core Methodologies for Bias Mitigation
Protocol for Competing Risk Analysis in HCC Prognostic Models
Competing risk bias represents a critical limitation in HCC prediction models, as traditional survival analyses overestimate HCC probability by ignoring the high rate of non-HCC mortality in cirrhosis patients [65].
- Experimental Rationale: To develop unbiased estimates of HCC risk by accounting for competing events, particularly non-HCC mortality.
- Materials:
- Clinical cohort with documented cirrhosis (e.g., patients with cured hepatitis C)
- Follow-up data including HCC incidence, non-HCC mortality, and study completion dates
- Standard prognostic factors (e.g., age, platelet count, albumin)
- Step-by-Step Procedure:
- Define Risk Sets: Establish follow-up time beginning at a consistent baseline (e.g., date of sustained virologic response achievement). Follow-up ends at HCC diagnosis, non-HCC death, or study completion [65].
- Model Development:
- Model 1 (Standard Cox Regression): Develop a prognostic model using standard Cox proportional hazards regression, ignoring competing risks.
- Model 2 (Fine-Gray Regression): Develop a comparable model using Fine-Gray regression, modeling the cumulative incidence of HCC directly while accounting for non-HCC mortality as a competing event [65].
- Statistical Analysis:
- Calculate absolute risk predictions for both models.
- Assess discrimination using Harrel's C-index for Model 1 and the Wolbers modified C-index for Model 2.
- Evaluate risk stratification agreement between models using percentile-based risk categories [65].
- Validation: Compare the mean predicted probabilities of HCC between models and assess the clinical impact of risk overestimation.
Protocol for EV-Derived lncRNA Biomarker Discovery with Limited Samples
Isolating and analyzing lncRNAs from extracellular vesicles (EVs) enables the discovery of highly specific biomarkers, but requires careful methodology to overcome sample size limitations.
- Experimental Rationale: To systematically identify HCC-associated lncRNA signatures from circulating EVs across disease progression stages.
- Materials:
- Serum or plasma samples from well-phenotyped patient cohorts (healthy controls, CHB, cirrhosis, HA, HCC)
- Size-exclusion chromatography columns (ES911, Echo Biotech)
- Ultrafiltration units (100kD)
- RNA Purification Kit (Simgen, 5202050)
- Transmission electron microscope, nanoparticle tracking analyzer, Western blot equipment
- Step-by-Step Procedure:
- Sample Preparation: Collect fasting venous blood in serum separator tubes or EDTA anticoagulant tubes. Process within 2 hours; centrifuge and store aliquots at -80°C [61].
- EV Isolation: Thaw samples and pre-filter through 0.8 μm filter. Separate via gel-permeation chromatography. Collect eluent from specific fractions (tubes 7-9) and concentrate using 100kD ultrafiltration [61].
- EV Characterization:
- Morphology: Use transmission electron microscopy with uranyl acetate staining.
- Size Distribution: Analyze by nanoparticle tracking analysis.
- Marker Validation: Confirm EV identity via Western blot for TSG101, Alix, CD9; confirm absence of calnexin [61].
- RNA Extraction & Sequencing: Extract total RNA from EVs using the purification kit with Buffer TL and Buffer EX. Perform high-throughput transcriptome sequencing [61].
- Bioinformatic Analysis:
- Identify differentially expressed lncRNAs across disease stages.
- Perform multi-step screening and time-series analysis to identify core lncRNAs associated with HCC progression.
- Construct lncRNA-miRNA-mRNA regulatory networks.
- Perform functional enrichment analysis (e.g., autophagy/MAPK pathways) and PPI network analysis to identify hub genes [61].
Protocol for Feature Reduction in Machine Learning Models
High-dimensional data from lncRNA studies necessitates feature reduction to prevent overfitting and enhance model performance, particularly with limited samples.
- Experimental Rationale: To optimize ML model performance by identifying the most relevant feature subset from high-dimensional lncRNA data.
- Materials:
- Normalized clinical and lncRNA expression dataset
- Computational environment with Python/R and necessary libraries (scikit-learn, etc.)
- Step-by-Step Procedure:
- Data Normalization: Preprocess data to standardize feature scales, improving model performance and convergence [66].
- Feature Reduction:
- Recursive Feature Elimination (RFE): Iteratively remove features, testing model performance at each iteration to identify the optimal feature subset [66].
- Principal Component Analysis (PCA): Transform the dataset into a set of linearly uncorrelated principal components to reduce dimensionality while preserving variance [66].
- Feature Optimization: Apply mutual information to rate feature importance for the classification task, optimizing the feature subset selection [66].
- Model Training & Validation: Apply multiple ML algorithms (Naive Bayes, SVM, Neural Networks, Decision Tree, KNN) to both original and reduced feature sets. Compare performance metrics (accuracy, precision, recall, F-score, execution time) [66].
Visual Workflows
EV-derived lncRNA Analysis Workflow
AI-Assisted HCC Screening Integration
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Materials for lncRNA Biomarker Studies
| Item Name | Manufacturer/Catalog Number | Function/Application | Key Consideration |
|---|---|---|---|
| Size-exclusion Chromatography Column | Echo Biotech / ES911 | Isolation of intact EVs from serum/plasma | Preserves EV integrity and biological activity [61] |
| Ultrafiltration Unit | Various / 100kD molecular weight cutoff | Concentration of EV samples post-isolation | Enables downstream molecular analyses [61] |
| RNA Purification Kit | Simgen / 5202050 | Extraction of high-quality total RNA from EVs | Optimized for low-concentration EV-derived RNA [61] |
| Antibody: TSG101 | Abcam / ab125011 | EV marker validation via Western blot | Confirms successful EV isolation [61] |
| Antibody: CD9 | Abcam / ab263019 | EV surface marker detection | Supports EV characterization and quantification [61] |
| Antibody: Calnexin | Proteintech / 10427-2-AP | Negative control for EV preparations | Confirms absence of cellular contaminants [61] |
| FujiFilm Laboratory Services | FujiFilm | Measurement of AFP, AFP-L3, and DCP | Standardized measurements for GALAD score calculation [69] |
| UniMatch AI Model | Custom development | Automated detection of liver lesions in ultrasound images | Reduces radiologist workload by 54.5% [68] |
| LivNet AI Model | Custom development | Classification of detected liver lesions | Improves specificity of HCC screening [68] |
The integration of machine learning with lncRNA biomarkers for HCC diagnosis requires meticulous attention to dataset limitations to ensure clinical applicability. The protocols and strategies outlined herein—including competing risk analysis, EV-derived lncRNA profiling, and strategic feature reduction—provide a methodological foundation for developing robust, generalizable models. Furthermore, AI-assisted screening integration demonstrates a viable path for implementing these models in clinical workflows while managing resource constraints. As the field advances, adherence to these rigorous methodological standards will be paramount for translating lncRNA biomarkers into clinically valuable tools that improve early HCC detection and patient outcomes.
The integration of machine learning (ML) with long non-coding RNA (lncRNA) biomarkers represents a transformative frontier in hepatocellular carcinoma (HCC) diagnostics. The clinical utility of these models hinges critically on their robustness and generalizability beyond the data on which they were trained. Model robustness ensures that diagnostic predictions remain accurate and reliable when applied to new patient cohorts, different sample types, or varying experimental conditions. Without proper validation frameworks, models risk overfitting—performing well on training data but failing in real-world clinical applications.
Cross-validation and hyperparameter tuning form the methodological bedrock for developing robust, clinically translatable models. These techniques are particularly crucial in HCC biomarker research due to the frequent challenges of limited sample sizes and high-dimensional data (where the number of features far exceeds the number of observations). For instance, studies analyzing lncRNA expression often work with dozens of biomarkers across hundreds of patients, creating a complex statistical landscape where proper validation is not just beneficial but essential for generating clinically meaningful results [70] [6] [14].
Cross-Validation Techniques for lncRNA Biomarker Models
Cross-validation (CV) provides a robust framework for estimating how ML models will generalize to independent datasets, making it indispensable for assessing the real-world performance of lncRNA-based HCC classifiers. The core principle involves partitioning data into subsets, training the model on some subsets, and validating it on the remaining subsets. This process is repeated multiple times to obtain a stable performance estimate.
Core Cross-Validation Methods
k-Fold Cross-Validation is the most widely adopted approach in HCC biomarker research. The dataset is randomly partitioned into k equal-sized folds. The model is trained on k-1 folds and validated on the remaining fold. This process is repeated k times, with each fold used exactly once as the validation set. The k results are then averaged to produce a single estimation. Studies in HCC diagnostics commonly employ 5-fold or 10-fold cross-validation, providing a reasonable balance between computational expense and performance estimation reliability [71] [14]. For example, in developing a model to differentiate HCC from controls using lncRNA profiles, 10-fold cross-validation demonstrated superior stability in performance metrics compared to single train-test splits [72].
Leave-One-Out Cross-Validation (LOOCV) represents an extreme form of k-fold CV where k equals the number of observations in the dataset. Each iteration uses a single sample as the validation set and all remaining samples as the training set. While computationally intensive, LOOCV is particularly valuable for small datasets where maximizing training data is crucial. This approach was effectively implemented in an HCC study combining multiple lncRNAs with conventional laboratory parameters, where it helped identify the most predictive biomarker combinations from limited patient samples [14].
Stratified k-Fold Cross-Validation maintains the same class distribution in each fold as in the complete dataset. This is particularly important for HCC biomarker studies where case-control ratios may be imbalanced. By preserving the proportion of HCC patients versus controls in each fold, stratified CV provides more reliable performance estimates for diagnostic models targeting early detection [71].
Nested Cross-Validation for Unbiased Performance Estimation
A critical advancement for avoiding optimistic bias in performance reporting is nested cross-validation (also known as double cross-validation). This approach implements two layers of cross-validation: an inner loop for hyperparameter tuning and an outer loop for performance estimation. This separation ensures that the test data in the outer loop never influences model development or parameter selection in the inner loop.
In HCC research, nested cross-validation was employed to validate a panel of 29 lncRNAs for predicting homologous recombination deficiency, where the dataset was divided into training (60%), validation (20%), and test (20%) sets using stratified sampling. The model was trained and tuned exclusively on the training set using 10-fold cross-validation, with final performance metrics evaluated on the completely held-out test set [73]. This rigorous approach provides realistic performance estimates for clinical translation.
Table 1: Comparison of Cross-Validation Techniques in HCC Biomarker Studies
| Technique | Key Characteristics | Best Use Cases | Reported Performance in HCC Studies |
|---|---|---|---|
| k-Fold CV | Divides data into k folds; trains on k-1, validates on 1; repeated k times | Medium to large datasets; standard model assessment | 5-fold and 10-fold CV commonly used; provides stable performance estimates [71] |
| Leave-One-Out CV | Each sample used once as validation; maximum training data | Small datasets (<100 samples); resource-intensive | Implemented in HCC RNA signature studies; computationally expensive but optimal for small samples [14] |
| Stratified k-Fold | Preserves class distribution in each fold | Imbalanced datasets (e.g., rare early-stage HCC) | Essential for maintaining HCC vs. control ratios; improves reliability [71] |
| Nested CV | Separates parameter tuning and performance estimation | Unbiased performance estimation; model selection | Used in lncRNA-HRD prediction; prevents optimistic bias in reported accuracy [73] |
Hyperparameter Tuning Methodologies
Hyperparameter tuning represents the systematic process of optimizing a model's configuration settings that are not learned directly from the data. For lncRNA-based HCC diagnostic models, appropriate hyperparameter selection can significantly enhance model performance and generalizability.
Fundamental Tuning Strategies
Grid Search represents the most straightforward approach, involving an exhaustive search across a predefined subset of hyperparameter space. Researchers specify a set of possible values for each hyperparameter, and the algorithm evaluates every possible combination. For example, when optimizing a Support Vector Machine (SVM) classifier for HCC detection using lncRNA expression profiles, a grid search might explore different kernel functions (linear, radial basis function, polynomial), regularization parameters (C values), and kernel-specific parameters (gamma, degree) [71] [72]. The main advantage is comprehensiveness—it doesn't miss the optimal combination within the specified range. However, computational demands grow exponentially with the number of hyperparameters, making it challenging for complex models or extensive search spaces.
Random Search differs by sampling hyperparameter combinations randomly from the specified distributions. Rather than exhaustively evaluating all possibilities, it sets a fixed number of iterations. Empirical studies have shown that random search often finds optimal or near-optimal configurations more efficiently than grid search, particularly when some hyperparameters have minimal impact on performance [71]. This approach is especially valuable during preliminary model development phases for HCC diagnostic models when computational resources are limited.
Bayesian Optimization represents a more sophisticated approach that builds a probabilistic model of the function mapping hyperparameters to model performance. It uses this model to select the most promising hyperparameters to evaluate in the next iteration. Bayesian optimization has demonstrated particular effectiveness for optimizing complex models like neural networks and gradient boosting machines, which have high-dimensional hyperparameter spaces and expensive evaluation times [14]. In one HCC study integrating multiple RNA biomarkers, Bayesian optimization achieved 98.75% accuracy in predicting HCC cases by efficiently navigating the complex parameter space of a LightGBM classifier [14].
Hyperparameter Tuning in Practice: HCC Case Examples
The practical implementation of hyperparameter tuning in HCC research varies by algorithm. For Random Forest classifiers commonly used in lncRNA biomarker studies, critical hyperparameters include the number of trees in the forest (nestimators), maximum depth of trees (maxdepth), minimum samples required to split a node (minsamplessplit), and minimum samples required at a leaf node (minsamplesleaf) [71] [72]. For Support Vector Machines, key parameters include the regularization parameter (C), kernel type, and kernel-specific parameters such as gamma for RBF kernels [71] [73].
Table 2: Key Hyperparameters for Common Algorithms in HCC Biomarker Research
| Algorithm | Critical Hyperparameters | Recommended Search Ranges | Impact on Model Performance |
|---|---|---|---|
| Random Forest | nestimators: 100-1000maxdepth: 5-50minsamplessplit: 2-10minsamplesleaf: 1-5 | Logarithmic scale for n_estimatorsLinear scale for depth and samples | Controls overfittingBalances bias-variance tradeoffAffects feature importance stability [71] |
| Support Vector Machine | C: 0.001-1000gamma: 0.0001-10kernel: linear, RBF, polynomial | Logarithmic scale for C and gammaDiscrete for kernel | Influences margin width and misclassification penaltyControls influence of individual samples [71] [73] |
| XGBoost | learningrate: 0.01-0.3maxdepth: 3-10subsample: 0.6-1.0colsample_bytree: 0.6-1.0 | Fine grid around default valuesLogarithmic for learning_rate | Affects convergence and overfittingControls row and column sampling [14] |
| Neural Networks | hiddenlayersizes: (10-500,) learning_rate: constant, adaptivealpha: 0.0001-0.1 | Varies significantly by architectureLogarithmic for regularization | Impacts model capacity and generalizationRegularization strength [71] |
Integrated Protocol for Robust HCC Diagnostic Model Development
This section provides a detailed, actionable protocol for developing and validating robust lncRNA-based HCC diagnostic models, integrating both cross-validation and hyperparameter tuning strategies.
Experimental Workflow for HCC Biomarker Model Validation
Step-by-Step Protocol
Step 1: Data Preparation and Partitioning
- Dataset Collection: Compile lncRNA expression data from relevant sources (e.g., GEO datasets, in-house RT-qPCR measurements). Include appropriate control samples (healthy liver, chronic hepatitis, cirrhosis) alongside HCC samples. Studies typically require samples from at least 50-100 patients per group for adequate power [70] [6].
- Quality Control: Remove samples with excessive missing data or outliers. For lncRNA expression data, apply normalization procedures such as DESeq2 for RNA-Seq data or the ΔΔCT method for qRT-PCR data [71] [25].
- Initial Partitioning: Perform an initial 80/20 split of the data into a training set (for model development and tuning) and a completely held-out test set (for final evaluation). Ensure stratification maintains the proportion of HCC cases and controls in both sets.
Step 2: Establish Nested Cross-Validation Framework
- Outer Loop Configuration: Implement k-fold cross-validation (typically k=5 or k=10) on the training set for performance estimation [71] [14].
- Inner Loop Configuration: Within each training fold of the outer loop, implement an additional j-fold cross-validation (typically j=5) specifically for hyperparameter tuning.
Step 3: Hyperparameter Optimization in Inner Loop
- Define Search Space: Specify the hyperparameter ranges to explore based on the selected algorithm (refer to Table 2 for guidance).
- Execute Search Method:
- Evaluation Metric: Select appropriate evaluation metrics for HCC diagnostics: area under the ROC curve (AUC), sensitivity, specificity, or balanced accuracy. For imbalanced datasets, consider F1-score or Matthews Correlation Coefficient [71].
- Identify Optimal Configuration: Select the hyperparameter set that maximizes the chosen metric across all inner loop validation folds.
Step 4: Model Training and Validation
- Train Final Model: Using the optimal hyperparameters identified in the inner loop, train the model on the complete training fold of the outer loop.
- Performance Assessment: Evaluate the model on the outer loop test fold, recording all performance metrics.
- Iteration: Repeat steps 2-4 for each fold in the outer loop.
Step 5: Final Model Evaluation and Reporting
- Aggregate Performance: Calculate mean and standard deviation of all performance metrics across the outer loop folds.
- Final Model Training: Train the model on the entire training set using the hyperparameter configuration that demonstrated the best average performance during nested CV.
- Independent Testing: Evaluate the final model on the completely held-out test set that was separated in Step 1.
- Model Interpretation: Analyze feature importance scores to identify the lncRNAs contributing most to HCC classification accuracy [6] [14].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Computational Tools for lncRNA Biomarker Studies
| Category | Specific Product/Tool | Application in HCC Biomarker Research | Key Features/Benefits |
|---|---|---|---|
| RNA Isolation | miRNeasy Mini Kit (QIAGEN) | Total RNA extraction from plasma/serum, tissue | Preserves lncRNA integrity; suitable for liquid biopsies [6] [25] |
| cDNA Synthesis | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) | Reverse transcription for lncRNA quantification | High-efficiency synthesis; compatible with challenging samples [6] |
| qRT-PCR | PowerTrack SYBR Green Master Mix (Applied Biosystems) | lncRNA expression quantification | Sensitive detection; compatible with high-throughput systems [6] |
| RNA Sequencing | Illumina HiSeq 2500/NovaSeq 6000 | Transcriptome-wide lncRNA profiling | Comprehensive lncRNA discovery; identifies novel isoforms [71] [72] |
| Data Analysis | R Studio with caret, mlr3 packages | Cross-validation and hyperparameter tuning | Unified interface for multiple ML algorithms; reproducible research [71] |
| ML Frameworks | Python Scikit-learn, XGBoost | Implementing classifiers and optimization | Comprehensive ML algorithms; efficient hyperparameter search [6] [14] |
The rigorous implementation of cross-validation and hyperparameter tuning methodologies is not merely a technical exercise but a fundamental requirement for developing clinically relevant lncRNA-based HCC diagnostic models. The integrated framework presented in this protocol ensures that performance estimates reflect true generalizability rather than over-optimistic results from overfitting. As the field advances toward liquid biopsy approaches and multi-analyte panels combining lncRNAs with other biomarker classes, these robustness assurance techniques will become increasingly critical for bridging the gap between research findings and clinical implementation.
Hepatocellular carcinoma (HCC) represents a global health challenge, ranking as the sixth most prevalent cancer worldwide and the fourth most common cause of cancer-related mortality [6]. The disease exhibits a particularly aggressive course, with a five-year survival rate that remains alarmingly low at 10-20% [74]. This poor prognosis is largely attributable to late diagnosis and the suboptimal efficacy of current therapies for advanced disease [74]. The established biomarker Alpha-fetoprotein (AFP) demonstrates significant limitations, with reported sensitivity ranging from 60-83% and specificity of 53-67% [6], while approximately 20-40% of HCC patients' tumor cells do not secrete AFP proteins at all [74]. These diagnostic shortcomings have intensified the search for more reliable biomarkers and created an urgent need for advanced analytical approaches that can integrate complex molecular data into clinically actionable insights.
Artificial intelligence (AI) and machine learning (ML) have emerged as transformative technologies in hepatology, demonstrating strong potential for diagnostic, prognostic, and workflow enhancement [75]. However, the clinical adoption of these advanced algorithms faces a significant barrier: their frequent characterization as "black boxes" whose decision-making processes remain opaque to clinicians and researchers [75]. This opacity creates justifiable skepticism in medical practice, where understanding the rationale behind a diagnosis or treatment recommendation is paramount for patient safety and trust. Explainable Artificial Intelligence (XAI) directly addresses this challenge by making the inner workings of complex models transparent and interpretable, thereby bridging the gap between algorithmic predictions and clinically actionable intelligence [74].
The integration of long non-coding RNA (lncRNA) biomarkers with XAI represents a particularly promising frontier in HCC research. lncRNAs, defined as non-coding RNAs greater than 200 nucleotides in length, play essential roles as regulators in physiological and pathological processes [6]. In HCC, they function as key regulators of oncogene and tumor suppressor gene expression, with differential expression patterns affecting cancer growth, survival, and therapeutic response [76]. The detection of HCC-associated lncRNAs in body fluids makes them particularly accessible for liquid biopsy approaches, highlighting their potential as valuable non-invasive biomarkers [6]. When combined with XAI methodologies, these molecular signatures can transition from mere correlative observations to comprehensible components of predictive models that clinicians can understand, trust, and ultimately apply in patient care decisions.
XAI Methodologies and Framework Implementation
Core XAI Algorithms and Their Mathematical Foundations
The development of clinically actionable XAI frameworks for HCC lncRNA integration relies on specific algorithmic approaches that balance predictive power with interpretability. Tree-based ensemble methods have demonstrated particular efficacy in this domain, with Extreme Gradient Boosting (XGBoost), Random Forest (RFC), and Extra Trees Classifiers (ETC) emerging as prominent models [74]. These algorithms learn the functional relationship (f) between molecular features (X) and clinical outcomes (Y) through iterative processes. In XGBoost, for instance, predictions are generated through an ensemble of sequentially trained trees, with each subsequent model focusing on the residuals (errors) of its predecessors [74]. Mathematically, this process can be represented as:
Ŷ = φ(X) = (1/n) ∑ₖ₌â‚â¿ fâ‚–(X)
where Ŷ represents the predictions, 1 ≤ k ≤ n, and n is the total number of functions learned by the n trees in the model [74]. The model's performance is optimized through a regularized objective function L(φ) that balances predictive accuracy with computational complexity:
L(φ) = ∑ᵢ l(ŷᵢ, yᵢ) + ∑ₖ Ω(fₖ)
where l is a differentiable convex loss function measuring differences between predictions (ŷᵢ) and actual targets (yᵢ), and Ω is a regularization term that penalizes model complexity to prevent overfitting [74]. This mathematical foundation provides both high predictive accuracy and a structured framework for subsequent interpretability analysis.
SHAP: A Unified Approach to Model Interpretability
To transform these sophisticated algorithms into clinically interpretable tools, researchers employ post-hoc explanation frameworks such as SHapley Additive exPlanations (SHAP) [74] [77]. SHAP operates on principles from cooperative game theory to quantify the marginal contribution of each feature (e.g., individual lncRNA expression levels) to the final prediction [77]. This approach provides a unified measure of feature importance that is consistent across different model architectures and aligns with clinical intuition by assigning each variable an importance value that represents its specific impact on an individual prediction.
The power of SHAP lies in its ability to generate both global interpretability (understanding the overall model behavior across the entire dataset) and local interpretability (understanding why a specific prediction was made for an individual patient) [77]. For HCC prognosis using lncRNA biomarkers, this means clinicians can both understand which biomarkers generally contribute most to accurate predictions and also see exactly which lncRNAs drove a specific prognostic assessment for their patient. This dual-level interpretability is crucial for building clinical trust and facilitating the integration of AI-driven insights into personalized treatment planning.
Table 1: XAI Algorithms for lncRNA Biomarker Integration in HCC
| Algorithm | Mechanism | Interpretability Strengths | Clinical Application |
|---|---|---|---|
| XGBoost | Gradient boosting with sequential tree building | High predictive accuracy with built-in regularization | Identification of non-linear relationships between lncRNA combinations |
| Random Forest | Bagging ensemble of decision trees | Natural feature importance metrics | Robust lncRNA signature discovery resistant to overfitting |
| SHAP | Game theory-based attribution values | Unified scale for feature importance across models | Translating model outputs to clinically understandable biomarker contributions |
Workflow for XAI Implementation in HCC Research
The practical implementation of XAI for lncRNA biomarker integration follows a structured workflow that transforms raw molecular data into clinically actionable insights. This process begins with data acquisition and preprocessing, followed by model training and validation, and culminates in the generation of interpretable outputs through explainability frameworks.
XAI Workflow for HCC lncRNA Integration
Experimental Protocols for XAI-Driven lncRNA Biomarker Research
Specimen Collection and RNA Isolation Protocol
The foundation of reliable XAI modeling in HCC lncRNA research begins with rigorous specimen collection and processing. For plasma-based liquid biopsy approaches, collect whole blood in EDTA-containing tubes from HCC patients and matched controls following standard phlebotomy procedures [6]. Process samples within 2 hours of collection through centrifugation at 1,500-2,000 × g for 10 minutes at 4°C to separate plasma, followed by a second centrifugation at 12,000 × g for 10 minutes to remove residual cellular debris [6]. Aliquot cleared plasma into RNase-free tubes and store at -80°C until RNA extraction.
For RNA isolation, use the miRNeasy Mini Kit or similar validated systems according to manufacturer's protocol with the following critical modifications for optimal lncRNA recovery [6]:
- Add 1 volume of plasma to 3 volumes of Qiazol lysis reagent
- Include synthetic spike-in controls for quality assessment
- Perform on-column DNase digestion for 15 minutes at room temperature
- Elute in 30-50 μL of RNase-free water after a 5-minute incubation
Quantify RNA yield and purity using spectrophotometry (A260/A280 ratio ≥1.8, A260/A230 ratio ≥2.0), and assess integrity through automated electrophoresis (RIN ≥7.0 for tissue samples; minimal fragmentation expected for plasma-derived RNA).
cDNA Synthesis and Quantitative RT-PCR
Reverse transcribe purified RNA into cDNA using the RevertAid First Strand cDNA Synthesis Kit with random hexamer primers according to manufacturer's protocol [6]. Use 100-500 ng of total RNA per 20 μL reaction, incubating at 25°C for 5 minutes, 42°C for 60 minutes, and 70°C for 5 minutes. Dilute synthesized cDNA 1:5 with nuclease-free water before qRT-PCR analysis.
For quantitative assessment of lncRNA expression, prepare reactions using PowerTrack SYBR Green Master Mix on a ViiA 7 real-time PCR system or equivalent platform [6]. Utilize primer sequences specifically designed for HCC-relevant lncRNAs:
Table 2: Primer Sequences for Key HCC-Associated lncRNAs
| lncRNA | Forward Primer (5'→3') | Reverse Primer (5'→3') | Amplicon Size | Clinical Significance |
|---|---|---|---|---|
| LINC00152 | CAGTGGAAAACCACCACCTG | GGCTGGACTTTCATTCCAAA | ~150 bp | Promotes cell proliferation through CCDN1 regulation; prognostic for shorter OS [6] [76] |
| GAS5 | GGCACTGAGATCCCTGGATT | TGGTGGTAGAGTGGCTGCTT | ~120 bp | Tumor suppressor; activates CHOP and caspase-9 apoptosis pathways [6] |
| UCA1 | Not specified in sources | Not specified in sources | - | Promotes HCC cell proliferation and apoptosis resistance [6] |
| LINC00853 | Not specified in sources | Not specified in sources | - | Potential diagnostic marker when combined with other lncRNAs [6] |
Perform all reactions in triplicate with the following cycling conditions: initial denaturation at 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Include non-template controls and inter-run calibrators to ensure technical reproducibility. Normalize expression data using the ΔΔCT method with GAPDH as the reference gene [6].
Data Integration and XAI Modeling Protocol
Integrate normalized lncRNA expression data with clinical parameters (e.g., AFP levels, liver function tests, demographic information) into a structured dataframe. For XAI model development, implement the following workflow using Python and Scikit-learn:
For model validation, employ comprehensive metrics including area under the receiver operating characteristic curve (AUC-ROC), precision-recall curves, calibration plots, and decision curve analysis to assess clinical utility [77]. The entire modeling process, from data loading through validation, typically requires minimal computational time, with studies reporting approximately 0.01-0.03 minutes for complete pipeline execution [74].
Performance Benchmarks and Clinical Validation
Diagnostic and Prognostic Performance of XAI-Integrated lncRNA Biomarkers
The implementation of XAI frameworks for lncRNA biomarker analysis has demonstrated remarkable performance improvements over conventional diagnostic approaches. Individual lncRNAs show moderate diagnostic accuracy when used alone, with sensitivity and specificity ranging from 60-83% and 53-67%, respectively [6]. However, when integrated through machine learning approaches, these biomarkers achieve substantially enhanced performance, with one study reporting 100% sensitivity and 97% specificity for HCC diagnosis [6].
For prognostic applications, specific lncRNA signatures have shown significant value in predicting clinical outcomes. The ratio of LINC00152 to GAS5 expression has been identified as a particularly powerful prognostic indicator, with higher ratios significantly correlating with increased mortality risk [6]. Numerous studies have validated the independent prognostic significance of individual lncRNAs through multivariate Cox proportional hazards regression analysis, confirming their value in predicting overall survival (OS) and recurrence-free survival (RFS) in HCC patients [76].
Table 3: Prognostic Performance of Key lncRNAs in HCC
| lncRNA | Expression in HCC | Hazard Ratio (95% CI) | P-value | Clinical Endpoint | Detection Method |
|---|---|---|---|---|---|
| LINC00152 | High | 2.524 (1.661-4.015) | 0.001 | Shorter OS | qRT-PCR [76] |
| LINC01146 | Low | 0.38 (0.16-0.92) | 0.033 | Longer OS | qRT-PCR [76] |
| LINC01554 | Low | 2.507 (1.153-2.832) | 0.017 | Shorter OS | qRT-PCR [76] |
| HOXC13-AS | High | 2.894 (1.183-4.223) | 0.015 | Shorter OS | qRT-PCR [76] |
| LASP1-AS | Low | 3.539 (2.698-6.030) | <0.0001 | Shorter OS | qRT-PCR [76] |
XAI-Driven Biomarker Discovery Beyond Conventional Approaches
Explainable AI approaches have facilitated the discovery of novel genetic biomarkers with prognostic significance that extend beyond traditional markers like AFP. Studies employing multi-model XAI frameworks have identified biomarkers such as TOP3B, SSBP3, and COX7A2L as consistently influential across multiple algorithms, suggesting their important role in improving predictive accuracy for HCC prognosis [74]. Notably, SSBP3 has been identified as a consistently influential gene across all AI models utilized, indicating its potential as a critical biomarker in HCC prognosis [74]. Similarly, COX7A2L has demonstrated significant influence in multiple models, further underscoring its possible importance in disease progression [74].
The composite application of these AI-identified biomarkers has been shown to markedly enhance prognostic accuracy beyond the capabilities of existing markers currently utilized in HCC detection and management [74]. This approach represents a paradigm shift from single-biomarker reliance to integrated molecular signatures that more comprehensively capture the biological complexity of hepatocellular carcinoma.
Successful implementation of XAI-driven lncRNA biomarker research requires access to specialized reagents, computational tools, and curated data resources. The following table summarizes essential components of the research toolkit for investigators in this field:
Table 4: Essential Research Resources for XAI-lncRNA Integration in HCC
| Resource Category | Specific Items | Function/Application | Example Products/Databases |
|---|---|---|---|
| Wet Lab Reagents | RNA Isolation Kit | Extraction of high-quality lncRNAs from plasma/tissue | miRNeasy Mini Kit (QIAGEN) [6] |
| cDNA Synthesis Kit | Reverse transcription for qRT-PCR analysis | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) [6] | |
| qPCR Master Mix | Quantitative measurement of lncRNA expression | PowerTrack SYBR Green Master Mix (Applied Biosystems) [6] | |
| Computational Tools | ML Libraries | Model development and training | Scikit-learn, XGBoost [74] [77] |
| Explainability Frameworks | Model interpretation and feature importance | SHAP (SHapley Additive exPlanations) [74] [77] | |
| Bioinformatics Platforms | Data preprocessing and analysis | Galaxy, DNAnexus [78] | |
| Data Resources | lncRNA Databases | Annotation and functional information | NONCODE, LNCipedia |
| HCC Omics Data | Model training and validation | HCCDB: Hepatocellular Carcinoma Expression Atlas [74] | |
| Biomarker Databases | Context for discovered biomarkers | MIRUMIR, exRNA Atlas [9] |
Pathway Visualization: lncRNA Mechanistic Roles in HCC Pathogenesis
The clinical utility of XAI-derived lncRNA biomarkers is enhanced by understanding their functional roles in HCC pathogenesis. lncRNAs participate in diverse molecular pathways that drive hepatocarcinogenesis through multiple mechanisms, including regulation of cell proliferation, apoptosis resistance, and metastatic potential.
lncRNA Functional Mechanisms in HCC
This pathway visualization illustrates how different lncRNA categories contribute to HCC pathogenesis through distinct molecular mechanisms. Oncogenic lncRNAs such as LINC00152 and UCA1 promote malignant phenotypes by enhancing cell cycle progression, proliferation signaling, and apoptosis evasion [6]. In contrast, tumor suppressor lncRNAs like GAS5 activate pathways that induce cell cycle arrest and apoptosis through CHOP and caspase-9 activation [6]. The detection of these differentially expressed lncRNAs in liquid biopsies provides the molecular basis for their utility as diagnostic, prognostic, and treatment response biomarkers when integrated with XAI analytical frameworks.
The application of explainable AI to these molecular pathways enables researchers and clinicians to move beyond simple correlative associations toward mechanistic understanding of how specific lncRNA expression patterns influence clinical outcomes. This integration of molecular biology with advanced analytics represents the future of precision oncology in hepatocellular carcinoma management.
Liquid biopsy represents a transformative approach in oncology, enabling non-invasive detection and monitoring of malignancies such as hepatocellular carcinoma (HCC) through the analysis of circulating biomarkers. Among these biomarkers, long non-coding RNAs (lncRNAs) have emerged as promising candidates due to their high cancer-specific expression and stability in biofluids [6] [79]. However, the quantification of lncRNAs from plasma presents significant technical challenges that hinder their clinical translation. This application note examines these hurdles within the broader context of integrating lncRNA biomarkers with machine learning (ML) for HCC diagnosis, providing detailed protocols and analytical frameworks to advance this promising field.
The pre-analytical, analytical, and post-analytical phases of lncRNA quantification introduce substantial variability. Key issues include inconsistent RNA recovery during isolation, amplification bias in detection methods, and lack of standardized normalization protocols [6] [25]. These technical barriers must be addressed to ensure the reproducible performance required for clinical application and effective ML model training.
Technical Hurdles in lncRNA Quantification
Pre-analytical Variability
Pre-analytical factors introduce significant variability in lncRNA quantification, potentially compromising downstream analysis and ML integration.
Blood Collection and Processing: The choice of anticoagulants in blood collection tubes (e.g., EDTA, citrate, heparin) can inhibit downstream enzymatic reactions during cDNA synthesis and PCR [25]. Plasma separation timing is critical; delays exceeding 2-4 hours can increase background RNA levels due to leukocyte lysis. Consistent centrifugation protocols (e.g., 704× g for 10 minutes for initial plasma separation, followed by higher-speed centrifugation to remove residual cells) are essential to minimize cellular RNA contamination [25].
Sample Storage Conditions: Repetitive freeze-thaw cycles can fragment lncRNAs and significantly alter quantification results. Studies store plasma samples at -70°C or lower to maintain RNA integrity for long-term storage [6] [25]. The development of standardized storage protocols across biobanks is necessary for multi-center studies.
Analytical Challenges
The analytical phase of lncRNA quantification presents hurdles in isolation, detection, and data normalization.
RNA Isolation Efficiency: The low abundance of lncRNAs in plasma and their coexistence with high concentrations of proteins and lipids complicate isolation. Commercial kits like the Norgen Biotek Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit or QIAGEN miRNeasy Mini Kit are commonly employed [6] [25]. However, varying extraction efficiencies between kits and batches can introduce significant technical variance, particularly for low-abundance targets.
Detection and Amplification Biases: Quantitative reverse-transcription PCR (qRT-PCR) remains the gold standard for lncRNA quantification due to its sensitivity, but it is susceptible to amplification bias [80] [6]. Factors such as primer specificity for the target lncRNA isoform, reverse transcriptase efficiency, and PCR inhibitor carryover from plasma can impact accuracy. Digital PCR offers potential for absolute quantification but requires further validation for lncRNA applications.
Normalization Strategies: The absence of universally stable reference genes in plasma represents a major hurdle for data normalization. Commonly used references include β-actin [25] and GAPDH [6], but their expression can vary under pathological conditions. Spike-in controls (e.g., synthetic non-human RNA sequences) are increasingly used to correct for technical variations in RNA isolation and reverse transcription efficiency, improving data robustness for ML analysis [6].
Post-analytical Complexities
Following data acquisition, standardization of analysis pipelines and data reporting is crucial.
Data Processing and QC Metrics: Establishing quality control thresholds for RNA purity (A260/A280 ratio), integrity, and the presence of genomic DNA contamination is essential. The inclusion of no-template controls and inter-plate calibrators in qRT-PCR runs helps identify contamination and technical drift [80] [25].
Standardization for ML Integration: For ML model development, consistent feature scaling and batch effect correction are required when merging datasets from different sources. Reporting standards must include detailed metadata on all pre-analytical and analytical steps to enable model reproducibility and external validation [80] [6].
Experimental Protocols for lncRNA Analysis
Protocol: Plasma Collection and RNA Isolation
Objective: To isolate high-quality total RNA from plasma for lncRNA quantification.
- Reagents and Equipment:
- Procedure:
- Blood Collection and Processing: Collect peripheral blood in EDTA tubes. Invert tubes gently to mix. Process within 2 hours of collection.
- Centrifuge at 704× g for 10 minutes at 4°C to separate plasma from cellular components [25].
- Carefully transfer the upper plasma layer to a new tube without disturbing the buffy coat.
- Perform a second centrifugation at 16,000× g for 10 minutes to remove any remaining cells or debris.
- Aliquot clarified plasma and store at -70°C if not used immediately.
- RNA Isolation: Use a commercial kit designed for low-concentration circulating RNA, following manufacturer instructions. The general workflow is:
- Add plasma (e.g., 500 μL) to a provided binding solution.
- Pass the mixture through an RNA-binding column.
- Wash columns with provided wash buffers to remove contaminants.
- Elute RNA in a small volume (e.g., 30-50 μL) of nuclease-free water.
- DNase Treatment: To eliminate genomic DNA contamination, treat purified RNA with DNase (e.g., Turbo DNase) according to the manufacturer's protocol [25].
- RNA Quality Assessment: Measure RNA concentration using a fluorometric method (e.g., Qubit) suitable for low-abundance RNA. Assess purity via spectrophotometry (A260/A280 ratio ~2.0 is ideal).
- Blood Collection and Processing: Collect peripheral blood in EDTA tubes. Invert tubes gently to mix. Process within 2 hours of collection.
Protocol: cDNA Synthesis and qRT-PCR
Objective: To convert isolated RNA to cDNA and quantify specific lncRNAs via qRT-PCR.
- Reagents and Equipment:
- Procedure:
- Reverse Transcription:
- Set up 20 μL reactions including RNA template, reverse transcriptase, random hexamers, dNTPs, and reaction buffer as per kit instructions.
- Use a thermal cycler with a standard program: 25°C for 10 min, 37°C for 120 min, 85°C for 5 min.
- Quantitative PCR:
- Prepare 10-20 μL reactions containing cDNA template, SYBR Green Master Mix, and forward and reverse primers.
- Run samples in triplicate alongside no-template controls and a standard curve if performing absolute quantification.
- Use the following cycling conditions on a real-time PCR system:
- Perform a melt curve analysis post-amplification to verify primer specificity.
- Data Analysis:
- Reverse Transcription:
Table 1: Example lncRNA Primers for HCC Research
| lncRNA | Primer Sequence (5' → 3') | Function / Relevance |
|---|---|---|
| LINC00152 | F: CTTACCGCGGCTCGAAATGGR: GAGCTGTTCCCACATCAGGC [80] | Oncogenic; promotes cell proliferation [6] [79] |
| UCA1 | Custom-designed by Thermo Fisher [6] | Oncogenic; role in proliferation and apoptosis [6] |
| GAS5 | Custom-designed by Thermo Fisher [6] | Tumor suppressor; induces apoptosis [6] |
| HULC | Sequence not specified in sources | Highly upregulated in liver cancer; oncogenic [25] |
| RP11-731F5.2 | Sequence not specified in sources | Potential biomarker for HCC risk and liver damage [25] |
Machine Learning Integration
The integration of lncRNA data with machine learning requires careful data curation and model selection to overcome technical noise and build robust diagnostic classifiers.
Data Preprocessing for ML
Feature Selection: ML algorithms like Random Forest (RF) and LASSO (Least Absolute Shrinkage and Selection Operator) regression are highly effective for identifying the most predictive lncRNAs from high-dimensional data. RF ranks features by importance based on Gini impurity, while LASSO penalizes the absolute size of regression coefficients, driving less important feature coefficients to zero [80]. These methods were successfully used to narrow down 55 differentially expressed lncRNAs to a panel of 5 key lncRNAs (NCAL1, CRNDE, HMGA1P4, EPIST, MT1JP) in colorectal cancer [80].
Data Normalization and Augmentation: Beyond traditional qPCR data normalization (2^(-ΔΔCq)), ML pipelines often apply z-score standardization or min-max scaling to ensure all features contribute equally to the model. For small datasets, techniques like synthetic minority over-sampling (SMOTE) can help balance classes and improve model generalizability.
ML Model Construction and Validation
Algorithm Selection: Support Vector Machines (SVM), Random Forest, and neural networks are frequently employed. For example, a study on HCC integrating four lncRNAs with conventional lab data used Scikit-learn in Python to build a model achieving 100% sensitivity and 97% specificity, far surpassing individual lncRNA performance [6].
Validation and Performance Metrics: Rigorous validation is critical. Models should be tested on held-out validation sets or through cross-validation. Performance is evaluated using Area Under the Curve (AUC) of ROC curves, sensitivity, specificity, and accuracy. An AUC > 0.7 is generally considered indicative of good diagnostic performance [80].
The following diagram illustrates the integrated workflow from sample processing to machine learning-based diagnosis.
Integrated lncRNA and ML Workflow for HCC Diagnosis
Performance Data and Validation
Robust validation is essential to demonstrate the clinical potential of lncRNA biomarkers and their performance in ML-driven diagnostic panels.
Table 2: Performance of lncRNA Biomarkers in HCC Detection
| lncRNA / Model | Sensitivity (%) | Specificity (%) | AUC | Sample Size (HCC/Control) | Notes |
|---|---|---|---|---|---|
| LINC00152 | 83 | 67 | >0.7 | 52/30 [6] | Individual performance |
| UCA1 | 60 | 53 | >0.7 | 52/30 [6] | Individual performance |
| GAS5 | 63 | 60 | >0.7 | 52/30 [6] | Individual performance |
| ML Model (4-lncRNA panel + lab data) | 100 | 97 | N/R | 52/30 [6] | Combined panel with machine learning |
| HULC | N/R | N/R | N/R | 41/22 [25] | Identified as a risk biomarker in CHC patients |
| RP11-731F5.2 | N/R | N/R | N/R | 41/22 [25] | Biomarker for HCC risk and liver damage |
The data in Table 2 highlights a critical finding: while individual lncRNAs show moderate diagnostic accuracy, their integration into a multi-marker panel and analysis with an ML model dramatically improves performance, achieving near-perfect sensitivity and specificity in one study [6]. This underscores the importance of combinatorial approaches and advanced computational analysis for effective HCC diagnosis.
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions
| Item | Function / Application | Example Products / Comments |
|---|---|---|
| Plasma RNA Kit | Isolation of high-quality circulating RNA from plasma/serum. | Norgen Biotek Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit; QIAGEN miRNeasy Mini Kit [6] [25] |
| DNase I | Removal of genomic DNA contamination from RNA preparations to prevent false-positive PCR results. | Turbo DNase (Thermo Fisher Scientific) [25] |
| Reverse Transcription Kit | Synthesis of complementary DNA (cDNA) from purified RNA templates. | High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher); RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) [80] [25] |
| SYBR Green Master Mix | Fluorescent dye for detection and quantification of PCR products in real-time qPCR. | Power SYBR Green PCR Master Mix (Thermo Fisher); PowerTrack SYBR Green Master Mix (Applied Biosystems) [80] [6] [25] |
| Reference Gene Primers | Essential control for normalizing lncRNA expression levels in qPCR. | Primers for β-actin or GAPDH [6] [25] (must be validated for stability in plasma) |
| lncRNA-specific Primers | Amplification and detection of target lncRNA sequences. | Designed using tools like Primer-BLAST; validated for specificity and efficiency [80] |
Standardizing the quantification of lncRNAs from plasma is a critical but surmountable challenge. By implementing rigorous protocols for pre-analytical processing, RNA isolation, and qRT-PCR, and by leveraging machine learning for data integration and analysis, researchers can overcome these technical hurdles. The remarkable diagnostic performance achieved by combining lncRNA panels with ML models, as demonstrated in recent HCC studies, provides a clear roadmap for the development of robust, non-invasive diagnostic tools. Future work must focus on the external validation of these integrated pipelines in large, multi-center cohorts to firmly establish their clinical utility.
Ethical and Privacy Considerations in AI-Driven Diagnostic Development
The integration of artificial intelligence (AI) and long non-coding RNA (lncRNA) biomarkers represents a transformative frontier in hepatocellular carcinoma (HCC) diagnostics. Machine learning models demonstrate exceptional capability in analyzing complex lncRNA expression patterns, achieving diagnostic accuracies surpassing traditional methods. For instance, one study integrating four lncRNAs (LINC00152, LINC00853, UCA1, and GAS5) with clinical parameters achieved 100% sensitivity and 97% specificity in HCC detection [6]. Similarly, random forest models utilizing minimal clinical predictors have reached 98.9% accuracy in detecting HCC [53]. However, this advanced diagnostic paradigm introduces significant ethical and privacy considerations that researchers must address throughout development and implementation. The collection and analysis of sensitive genomic data within AI systems necessitates robust frameworks to maintain patient confidentiality while advancing diagnostic innovation.
Foundational Research: AI-Enhanced lncRNA Biomarkers for HCC
Diagnostic Performance of Circulating lncRNAs in HCC
Long non-coding RNAs have emerged as promising liquid biopsy biomarkers due to their remarkable stability in circulation and specific dysregulation in hepatocellular carcinoma. Their resistance to nuclease-mediated degradation and presence in various biofluids make them ideal candidates for non-invasive diagnostics [81]. Numerous studies have validated the diagnostic potential of specific lncRNAs, both individually and as combined signatures.
Table 1: Diagnostic Performance of Key lncRNAs in Hepatocellular Carcinoma
| lncRNA Biomarker | Sample Type | Sensitivity (%) | Specificity (%) | AUC | Citation |
|---|---|---|---|---|---|
| LINC00152 | Plasma | 83 | 67 | 0.78 | [6] |
| UCA1 | Serum | 82 | 82 | - | [81] |
| GAS5 | Plasma | 60 | 53 | - | [6] |
| LINC00853 | Plasma | 63 | 67 | - | [6] |
| Four-lncRNA Panel (ML Model) | Plasma | 100 | 97 | - | [6] |
| MALAT1 | Plasma | - | 85 | - | [81] |
| HULC | Blood | - | - | - | [81] |
AI Model Performance in HCC Detection
Machine learning algorithms significantly enhance the diagnostic utility of lncRNA biomarkers by integrating them with clinical parameters to create powerful predictive models. These approaches outperform conventional statistical methods in detecting complex, non-linear patterns within multi-dimensional data.
Table 2: Performance of AI Models in HCC Detection Using Biomarkers and Clinical Data
| AI Model | Features Utilized | Sensitivity (%) | Specificity (%) | Accuracy (%) | AUC | Citation |
|---|---|---|---|---|---|---|
| Support Vector Machine | 22 clinical variables, CTCs, CECs | 100.0 | 98.7 | 98.7 | 0.971 | [82] |
| Random Forest | 7 clinical predictors | 90.5 | 99.8 | 98.9 | 0.999 | [53] |
| LightGBM | 7 clinical predictors | 94.9 | 99.5 | 99.1 | 0.999 | [53] |
| Custom ML Model | 4 lncRNAs + laboratory parameters | 100.0 | 97.0 | - | - | [6] |
| AI Pipeline (Strategy 4) | Ultrasound imaging | 95.6 | 78.7 | - | 0.872 | [68] |
| Blood-based AI Model | Routine blood tests | 80.0 | 81.0 | - | 0.894 | [32] |
Experimental Protocols: lncRNA Quantification and AI Integration
Protocol 1: Plasma lncRNA Quantification and Analysis
Objective: Isolate and quantify circulating lncRNAs from patient plasma samples for HCC diagnostic development.
Materials and Reagents:
- miRNeasy Mini Kit (QIAGEN, cat no. 217004) for RNA isolation
- RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, cat no. K1622) for reverse transcription
- PowerTrack SYBR Green Master Mix (Applied Biosystems, cat no. A46012) for qRT-PCR
- Primers for target lncRNAs (LINC00152, LINC00853, UCA1, GAS5) and housekeeping gene GAPDH
- ViiA 7 real-time PCR system (Applied Biosystems)
Methodology:
- Sample Collection and Processing: Collect whole blood in EDTA-containing tubes. Process within 2 hours of collection with centrifugation at 2,000 × g for 10 minutes at 4°C. Transfer plasma to clean tubes and store at -80°C until RNA extraction [6].
RNA Isolation: Use miRNeasy Mini Kit according to manufacturer's protocol. Add appropriate volumes of QIAzol Lysis Reagent to plasma samples. Add chloroform and separate phases by centrifugation. Transfer aqueous phase to new collection tubes and mix with ethanol. Transfer to RNeasy Mini spin columns and wash with buffer solutions. Elute RNA in RNase-free water [6].
cDNA Synthesis: Perform reverse transcription using RevertAid First Strand cDNA Synthesis Kit with 1μg of total RNA input in 20μL reaction volume. Use thermal cycler program: 25°C for 5 minutes, 42°C for 60 minutes, 70°C for 5 minutes [6].
Quantitative RT-PCR: Prepare reactions with PowerTrack SYBR Green Master Mix. Use standard cycling conditions: 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Perform all reactions in triplicate. Calculate relative expression using the ΔΔCT method with GAPDH as reference gene [6].
Data Analysis: Normalize expression levels to reference gene. Determine optimal cutoff values using receiver operating characteristic (ROC) curve analysis. Calculate sensitivity, specificity, and area under the curve (AUC) for diagnostic accuracy assessment.
Protocol 2: Machine Learning Model Development for HCC Diagnosis
Objective: Develop and validate a machine learning model integrating lncRNA expression data with clinical parameters for HCC diagnosis.
Materials and Software:
- Python programming language with Scikit-learn library
- Clinical dataset including demographic, laboratory, and lncRNA expression values
- Computing environment with adequate processing power (minimum 8GB RAM)
Methodology:
- Data Preprocessing:
- Compile comprehensive dataset including lncRNA expression levels (LINC00152, LINC00853, UCA1, GAS5), standard laboratory values (ALT, AST, AFP, total bilirubin, albumin), and demographic information [6].
- Handle missing data using appropriate imputation methods (e.g., k-nearest neighbors imputation).
- Normalize continuous variables using z-score standardization to ensure equal weighting in model training.
- Categorize categorical variables using one-hot encoding.
Feature Selection:
- Apply multiple feature selection techniques including recursive feature elimination with cross-validation, random forest feature importance, and Lasso regression [53].
- Identify optimal feature set balancing model performance and complexity.
- Validate feature selection through domain expert consultation to ensure clinical relevance.
Model Training:
- Split dataset into training (80%) and testing (20%) sets using stratified sampling to maintain class distribution.
- Train multiple machine learning algorithms including logistic regression, support vector machines, random forests, and gradient boosting machines [53].
- Implement hyperparameter tuning using grid search or random search with cross-validation.
- Employ k-fold cross-validation (typically k=5 or k=10) to reduce overfitting and validate model stability.
Model Validation:
- Evaluate model performance on held-out test set using metrics including accuracy, sensitivity, specificity, AUC-ROC, and F1-score.
- Perform internal validation through bootstrapping techniques to assess model calibration.
- Conduct external validation when possible using independent cohort data to evaluate generalizability [82].
- Compare model performance against traditional diagnostic approaches (e.g., AFP alone) using DeLong's test for AUC comparisons.
Diagram 1: HCC diagnostic development workflow integrating ethical safeguards.
Ethical Considerations in AI-Driven lncRNA Diagnostic Development
Data Privacy and Genomic Information Protection
The development of AI models for HCC diagnosis utilizing lncRNA biomarkers requires extensive genomic and clinical data, creating significant privacy challenges. lncRNA expression data constitutes sensitive health information that could potentially reveal insights about disease predisposition beyond HCC. Researchers must implement comprehensive data protection strategies including:
De-identification Protocols: Implement rigorous de-identification procedures that remove all 18 HIPAA-defined personal identifiers from genomic and clinical data. However, complete anonymization of genomic data remains challenging due to the inherent identifiability of genetic information [83].
Secure Data Storage: Utilize encrypted databases with access controls based on role-based permissions. Implement audit trails to monitor data access and modification. Consider federated learning approaches that allow model training without transferring raw patient data between institutions [83].
Data Minimization: Collect only lncRNA and clinical data elements essential for the diagnostic model development. Establish data retention policies that specify appropriate timelines for data destruction once analytical purposes are fulfilled [9].
Algorithmic Bias and Fairness
Machine learning models may perpetuate or amplify existing healthcare disparities if trained on non-representative datasets. This concern is particularly relevant for HCC diagnostic models given the varying lncRNA expression patterns across different ethnic populations [53].
Representative Recruitment: Ensure study populations include diverse demographic representation, particularly encompassing ethnic groups with high HCC prevalence such as Asian and African populations [53] [81].
Bias Assessment: Implement rigorous testing for algorithmic bias across different subpopulations using fairness metrics such as demographic parity, equality of opportunity, and predictive value parity [9].
Model Transparency: Document limitations of trained models regarding population subgroups where performance may be degraded. Provide clear guidance on appropriate use populations in clinical implementation [83].
Informed Consent in Genomic Research
The complex nature of AI-driven lncRNA research necessitates enhanced informed consent processes that address specific challenges of genomic data and artificial intelligence applications.
Comprehensibility: Develop consent materials that explain lncRNA biomarkers, AI methodologies, and potential implications in accessible language without scientific jargon.
Future Use Specificity: Clearly specify potential future research applications of collected genomic data and provide tiered consent options when possible [9].
Withdrawal Procedures: Establish straightforward procedures for participants to withdraw from studies, including protocols for data destruction when feasible [84].
Privacy-Preserving Protocols for lncRNA Data Handling
Protocol 3: Ethical Data Collection and Anonymization
Objective: Establish guidelines for ethical collection and processing of lncRNA data that preserves participant privacy while maintaining data utility for AI model development.
Materials:
- Unique subject identification system
- Secure, encrypted database
- Data encryption software
Methodology:
- Informed Consent Process:
- Obtain institutional review board (IRB) approval before study initiation.
- Develop comprehensive consent forms detailing specific lncRNA biomarkers to be analyzed, planned AI methodologies, potential future research uses, and data sharing parameters.
- Include explicit provisions regarding the handling of incidental findings from lncRNA analysis.
Data De-identification:
- Replace direct identifiers (name, medical record number, etc.) with randomly generated subject codes.
- Maintain separate linkage files connecting subject codes to identifiers in encrypted, password-protected files with limited access.
- Remove all elements not essential for analysis (exact dates, geographic details beyond region) while preserving data utility through relative dating or age ranges.
Data Security Measures:
- Implement role-based access controls with minimum necessary access principles.
- Utilize end-to-end encryption for data transfer between institutions.
- Store genomic data in format-specific encrypted containers with audit trails logging all access attempts.
Diagram 2: Privacy-preserving data flow for AI-driven lncRNA research.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Materials for lncRNA Biomarker Development
| Reagent/Material | Manufacturer | Function | Application in Protocol |
|---|---|---|---|
| miRNeasy Mini Kit | QIAGEN (cat no. 217004) | RNA isolation from plasma samples | Total RNA extraction including small and long non-coding RNAs [6] |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific (cat no. K1622) | Reverse transcription | cDNA synthesis from RNA templates for qRT-PCR analysis [6] |
| PowerTrack SYBR Green Master Mix | Applied Biosystems (cat no. A46012) | Quantitative PCR | Detection and quantification of specific lncRNA targets [6] |
| ViiA 7 Real-Time PCR System | Applied Biosystems | Amplification and detection | Precise quantification of lncRNA expression levels [6] |
| Custom lncRNA Primers | Thermo Fisher Scientific | Target amplification | Specific detection of LINC00152, LINC00853, UCA1, GAS5 [6] |
| Python Scikit-learn Library | Open Source | Machine learning implementation | Model development and validation [6] [53] |
The integration of AI and lncRNA biomarkers for HCC diagnosis represents a promising diagnostic advancement with demonstrated exceptional performance in preliminary studies. However, responsible development requires parallel attention to the significant ethical and privacy considerations inherent in handling sensitive genomic data. By implementing robust privacy-preserving protocols, ensuring algorithmic fairness, maintaining transparency in AI methodologies, and establishing comprehensive ethical frameworks, researchers can advance this promising diagnostic paradigm while upholding the highest standards of research ethics and patient protection. The future of AI-driven HCC diagnostics depends not only on technical excellence but also on maintaining patient trust through ethical rigor.
Proving Efficacy: Validation Frameworks, Performance Metrics, and Comparative Analysis
Within the broader thesis on the machine learning (ML) integration of long non-coding RNA (lncRNA) biomarkers for Hepatocellular Carcinoma (HCC) diagnosis, the rigorous benchmarking of performance metrics is a critical step. Sensitivity, specificity, and the Area Under the Receiver Operating Characteristic Curve (AUC-ROC) provide the fundamental quantitative framework for evaluating the clinical potential and diagnostic accuracy of these novel biomarker panels [54]. This document outlines standardized protocols for performing this essential benchmarking analysis, synthesizing methodologies from recent peer-reviewed studies to create a cohesive application note for researchers, scientists, and drug development professionals.
Performance Benchmarking of lncRNA Signatures and ML Models
The diagnostic performance of individual lncRNAs, multi-lncRNA signatures, and ML models integrating diverse data types has been quantitatively assessed in recent literature. The table below summarizes key quantitative benchmarks from contemporary studies.
Table 1: Performance Benchmarks of lncRNA-Based Diagnostic Approaches for HCC
| Biomarker / Model | Sensitivity (%) | Specificity (%) | AUC-ROC | Clinical Context / Notes | Source |
|---|---|---|---|---|---|
| 3-lncRNA Disulfidptosis Signature | Not Specified | Not Specified | 0.756 (1-year), 0.695 (3-year), 0.701 (5-year) | Prognostic prediction of overall survival | [85] |
| Individual lncRNAs (LINC00152, UCA1, etc.) | 60 - 83 | 53 - 67 | Moderate individual accuracy | Diagnostic; performance improved in panels | [6] |
| ML Model (LncRNAs + Clinical Vars) | 100 | 97 | ~0.99 (inferred) | Diagnostic; integrates lncRNAs with standard lab tests | [6] |
| LGBM Model (RNA Signature Panel) | Accuracy: 98.75% | Accuracy: 98.75% | Not Specified | Diagnostic; model includes mRNAs, miRNAs, and lncRNAs | [14] |
| 4-lncRNA Early Recurrence Signature | Not Specified | Not Specified | High (exact value not specified) | Prognostic; predictive performance enhanced when combined with AFP and TNM stage | [15] |
Experimental Protocols for Benchmarking Analysis
Protocol: Establishing the Gold Standard and Patient Cohort
The accuracy of any benchmarking effort is contingent on a robust and unambiguous definition of the ground truth.
- Objective: To define the patient cohorts and diagnostic criteria that will serve as the reference standard for evaluating the lncRNA biomarker.
- Materials: Patient clinical records, imaging data (Ultrasound, CT, MRI), histopathology reports, and serum biomarker levels (e.g., AFP).
- Procedure:
- Cohort Definition: Recruit a cohort of subjects that includes:
- HCC Patients: Diagnosis confirmed via histopathological examination of tissue biopsy or non-invasive imaging criteria per established systems like LI-RADS [54].
- Control Groups: Age-matched healthy controls and patients with benign liver conditions (e.g., chronic hepatitis, cirrhosis) to assess specificity [14].
- Data Annotation: For each subject, compile definitive classification (HCC vs. control) based on the reference standard. For prognostic studies (e.g., early recurrence), clearly define the endpoint (e.g., recurrence within 24 months post-surgery) and annotate patient outcomes during follow-up [15].
- Cohort Splitting: Randomly divide the cohort into a training set (e.g., ~70%) for model/signature development and a validation set (e.g., ~30%) for unbiased performance benchmarking [15].
- Cohort Definition: Recruit a cohort of subjects that includes:
Protocol: qRT-PCR Validation of lncRNA Biomarkers
The quantitative reverse transcription polymerase chain reaction (qRT-PCR) is the gold standard for validating lncRNA expression levels.
- Objective: To accurately quantify the expression levels of candidate lncRNAs in patient serum or plasma samples.
- Research Reagent Solutions:
- miRNeasy Mini Kit (Qiagen): For purification of total RNA, including small RNAs, from serum/plasma [14].
- RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific): For reverse transcription of RNA into stable cDNA [6].
- PowerTrack SYBR Green Master Mix (Applied Biosystems): For fluorescent-based detection of amplified DNA during qRT-PCR [6].
- Primer Sets: Specific oligonucleotide primers designed for target lncRNAs and reference genes (e.g., GAPDH, RNA18S) [86] [6].
- Procedure:
- Sample Collection & RNA Extraction: Collect peripheral blood in EDTA tubes and isolate plasma via centrifugation. Extract total RNA using a commercial kit according to the manufacturer's protocol [14].
- Reverse Transcription: Synthesize cDNA from a standardized amount of total RNA using a reverse transcriptase kit.
- Quantitative PCR: Perform qRT-PCR reactions in duplicate or triplicate for each sample. The reaction mix typically includes cDNA template, SYBR Green Master Mix, and forward/reverse primers.
- Data Analysis: Calculate the relative expression of each lncRNA using the comparative 2^(-ΔΔCt) method, normalizing to the expression of a stable reference gene [86] [6] [14].
Protocol: Statistical Analysis and Metric Calculation
This protocol details the computation of key performance metrics from the experimental data.
- Objective: To calculate sensitivity, specificity, and AUC-ROC for lncRNA biomarkers or derived models.
- Materials: Statistical software (e.g., R, SPSS, Python with scikit-learn), expression data, and clinical classifications.
- Procedure:
- Risk Score Calculation (for multi-lncRNA signatures): For prognostic or diagnostic signatures, calculate a risk score for each patient. This is often derived from a Cox regression or other multivariate model. The formula is typically:
Risk Score = Σ (Coefficient_i × Expression_i)for each lncRNA in the signature [85] [15]. - ROC Curve Generation: Use statistical software (e.g., the
pROCpackage in R) to generate the ROC curve. The lncRNA expression level (or the risk score) is used as the predictor variable, and the clinical diagnosis (HCC vs. control) is used as the outcome variable [85] [86] [15]. - AUC Calculation: The software will compute the AUC, which represents the probability that the model will rank a randomly chosen positive instance higher than a randomly chosen negative instance.
- Determination of Sensitivity/Specificity:
- Identify the optimal cut-off value on the ROC curve. This is often the point that maximizes the Youden's Index (Sensitivity + Specificity - 1).
- Create a confusion matrix based on this cut-off.
- Calculate metrics:
- Risk Score Calculation (for multi-lncRNA signatures): For prognostic or diagnostic signatures, calculate a risk score for each patient. This is often derived from a Cox regression or other multivariate model. The formula is typically:
Workflow Visualization
The following diagram illustrates the integrated workflow for benchmarking lncRNA biomarkers, from sample collection to clinical application.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Kits for lncRNA Biomarker Research
| Item | Function / Application | Example Product / Note |
|---|---|---|
| RNA Extraction Kit | Purification of total RNA (including lncRNAs) from serum, plasma, or tissues. Critical for sample integrity. | miRNeasy Mini Kit (Qiagen) [14] |
| cDNA Synthesis Kit | Reverse transcription of RNA to stable complementary DNA (cDNA) for downstream PCR applications. | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) [6] |
| qRT-PCR Master Mix | Fluorescent-based detection for accurate quantification of lncRNA expression levels. | PowerTrack SYBR Green Master Mix (Applied Biosystems) [6] |
| Primer Sets | Specific oligonucleotides designed to amplify target lncRNAs and reference genes for normalization. | Custom LNA-enhanced primers can improve specificity [14] |
| Statistical Software | For ROC/AUC analysis, survival analysis, and machine learning model construction. | R packages: pROC, survival, glmnet; Python: scikit-learn [85] [86] [15] |
The clinical translation of long non-coding RNA (lncRNA) biomarkers for hepatocellular carcinoma (HCC) requires robust validation strategies that extend beyond initial discovery cohorts. External validation through independent cohort studies and public dataset verification represents a critical step in establishing prognostic and diagnostic reliability, ensuring that developed signatures generalize across diverse populations and experimental conditions. This verification process is particularly crucial for machine learning-based lncRNA models, which must demonstrate stability and reproducibility before clinical implementation [87] [37]. The integration of multiple validation approaches strengthens the evidence base for lncRNA biomarkers, separating truly robust signatures from those that may be overfitted to specific datasets or patient populations.
Within HCC research, external validation has revealed significant insights into disease progression and therapeutic response. For instance, multiple studies have demonstrated that lncRNA signatures not only predict overall survival but also correlate with immune infiltration patterns and drug sensitivity, providing a more comprehensive understanding of their clinical utility [87] [88] [89]. The emergence of public genomic data repositories has significantly accelerated this validation process, enabling researchers to test biomarker performance across geographically distinct populations with varied etiological risk factors including HBV, HCV, and non-alcoholic fatty liver disease.
Framework for External Validation of lncRNA Biomarkers
Core Components of a Comprehensive Validation Strategy
Table 1: Key Components of External Validation Strategies for lncRNA Biomarkers in HCC
| Validation Component | Description | Common Data Sources | Key Performance Metrics |
|---|---|---|---|
| Independent Cohort Validation | Testing biomarker performance in a completely separate patient population from the training set | ICGC, in-house clinical cohorts, multi-institutional collaborations | Overall survival prediction, disease-free survival, diagnostic accuracy |
| Temporal Validation | Assessing biomarker performance in samples collected during different time periods | Prospective cohort studies, biobanks | Sensitivity, specificity, AUC stability over time |
| Geographical Validation | Verifying biomarker efficacy across diverse ethnic and regional populations | International consortia, multi-center studies | Consistency of hazard ratios, predictive accuracy across subgroups |
| Methodological Validation | Confirming results across different technical platforms and protocols | Cross-platform comparisons (RNA-seq, qPCR, microarrays) | Technical reproducibility, concordance between measurement methods |
| Clinical Context Validation | Evaluating biomarker performance in specific clinical scenarios (early detection, recurrence prediction) | Disease-specific cohorts (e.g., HBV-related HCC, early-stage HCC) | Clinical utility metrics, decision curve analysis |
A robust external validation framework for lncRNA biomarkers in HCC incorporates multiple complementary approaches. Independent cohort validation remains the foundation, requiring testing in populations completely separate from the discovery cohort to prevent overfitting [87] [37]. Temporal validation ensures that biomarker performance remains consistent across different time periods, addressing potential cohort-specific effects. Geographical validation is particularly important for HCC given the varying etiological factors across regions, with HBV predominating in some areas and HCV or NAFLD in others [11] [25]. Methodological validation confirms that lncRNA signatures perform consistently across different measurement platforms, while clinical context validation establishes utility for specific applications such as early detection or recurrence prediction.
The workflow for external validation typically progresses from computational analyses using public datasets to experimental confirmation. As demonstrated in multiple studies, the process begins with validation in independent public cohorts such as TCGA-LIHC or ICGC, followed by technical validation using RT-qPCR in local or multi-center cohorts, and culminates in functional studies to establish biological plausibility [87] [37] [89]. This sequential approach ensures that only the most promising biomarkers advance to resource-intensive experimental stages.
Public Genomic Data Repositories for HCC Research
Table 2: Public Data Repositories for External Validation of HCC lncRNA Biomarkers
| Database | Primary Content | Sample Characteristics | Validation Applications |
|---|---|---|---|
| The Cancer Genome Atlas (TCGA-LIHC) | Multi-omics data including RNA-seq, clinical information, survival data | ~374 HCC samples, 50 normal adjacent tissues [87] | Prognostic signature validation, molecular subtyping, survival analysis |
| International Cancer Genome Consortium (ICGC) | Genomic, transcriptomic, epigenomic data from international cohorts | 231 HCC samples with clinical prognostic characteristics [87] | Independent prognostic validation, cross-population generalizability |
| Gene Expression Omnibus (GEO) | Curated microarray and high-throughput sequencing data | Multiple HCC datasets with varying clinical annotations | Technical validation across platforms, meta-analyses |
| Genomics of Drug Sensitivity in Cancer (GDSC) | Drug response data and genomic profiles | Pharmacogenomic data for anticancer compounds | Drug sensitivity prediction validation [87] [89] |
Public data repositories provide invaluable resources for external validation of lncRNA biomarkers in HCC. TCGA-LIHC serves as a primary source for discovery and initial validation, containing comprehensive molecular profiling data alongside detailed clinical annotations [87] [37]. The ICGC offers independently generated datasets that enable validation across different populations and sequencing platforms. These repositories collectively enable researchers to assess whether lncRNA signatures maintain predictive power across different patient populations, technical platforms, and clinical contexts, providing essential evidence for generalizability before proceeding to costly prospective validation studies.
Experimental Protocols for External Validation
Protocol 1: Computational Validation Using Public Datasets
Objective: To validate the prognostic performance of lncRNA signatures using independent public genomic datasets.
Materials and Reagents:
- R or Python programming environments with necessary bioinformatics packages
- Public dataset access (TCGA, ICGC, GEO)
- Clinical annotation files for the validation cohorts
Procedure:
- Data Acquisition and Preprocessing: Download RNA-seq data and corresponding clinical information for the validation cohort (e.g., ICGC, n=231 HCC samples) [87]. Normalize expression data using the same method applied in the discovery phase (e.g., FPKM, TPM).
- Signature Score Calculation: Apply the previously established lncRNA signature algorithm to the validation dataset. For a multivariable signature, calculate risk scores using the published formula: Risk score = Σ(Expressionl ncRNA × Coefficientl ncRNA) [37] [89]
- Stratification and Survival Analysis: Divide patients into high-risk and low-risk groups based on the optimal cutoff value determined in the training set or using the validation cohort's median risk score. Perform Kaplan-Meier survival analysis with log-rank tests to compare overall survival (OS) and disease-free survival (DFS) between groups [87] [37].
- Performance Metrics Calculation: Evaluate signature performance using:
- Time-dependent receiver operating characteristic (ROC) analysis at 1, 3, and 5 years
- Concordance index (C-index) for prognostic accuracy
- Hazard ratios (HR) with confidence intervals from Cox regression models
- Clinical Utility Assessment: Conduct univariate and multivariate Cox regression analyses to determine whether the lncRNA signature provides prognostic information independent of established clinical factors such as TNM stage, AFP level, and vascular invasion [37].
Troubleshooting Tips:
- Address batch effects between discovery and validation datasets using combat or other normalization methods
- Ensure consistent lncRNA annotation across different genomic builds
- Verify that clinical endpoints (e.g., overall survival, recurrence) are defined consistently across cohorts
Protocol 2: Experimental Validation in Independent Clinical Cohorts
Objective: To technically validate lncRNA biomarker expression patterns in an independent clinical cohort using quantitative PCR.
Materials and Reagents:
- Plasma/serum samples from independent HCC cohort and appropriate controls
- RNA isolation kit (e.g., Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit) [25]
- DNase treatment reagents (e.g., Turbo DNase)
- cDNA synthesis kit (e.g., High-Capacity cDNA Reverse Transcription Kit)
- Power SYBR Green PCR Master Mix
- Quantitative PCR system (e.g., StepOne Plus System)
- Primers for target lncRNAs and reference genes
Procedure:
- Cohort Design and Sample Collection: Establish a clearly defined independent validation cohort with appropriate sample size calculation. Include HCC patients and relevant controls (chronic liver disease, healthy controls) matched for key clinical parameters [6] [25]. Obtain ethical approval and informed consent.
- RNA Extraction: Isolate total RNA from 500 μL plasma/serum using specialized kits for liquid biopsy samples. Include DNase treatment step to remove genomic DNA contamination [25].
- cDNA Synthesis and qPCR: Reverse transcribe RNA to cDNA using validated kits. Perform quantitative PCR with Power SYBR Green chemistry under the following conditions: initial denaturation at 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 62°C for 1 minute [25].
- Data Normalization and Analysis: Calculate relative expression using the 2^(-ΔΔCt) method with β-actin or GAPDH as reference genes [6] [25]. Verify assay specificity through dissociation curve analysis.
- Statistical Validation: Assess diagnostic performance using ROC curve analysis. Evaluate correlation with clinical parameters using appropriate statistical tests (Pearson correlation, Mann-Whitney U test, etc.) [25].
Troubleshooting Tips:
- Include no-template controls to detect contamination
- Analyze samples in triplicate to ensure technical reproducibility
- Use standardized sample collection and processing protocols to minimize pre-analytical variability
Visualization of Validation Workflows and Analytical Frameworks
Diagram 1: Integrated workflow for external validation of lncRNA biomarkers in HCC, combining computational approaches with experimental confirmation.
Table 3: Essential Research Reagents for lncRNA Biomarker Validation in HCC
| Category | Specific Product/Kit | Manufacturer | Application Note |
|---|---|---|---|
| RNA Isolation | Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit | Norgen Biotek | Optimized for low-abundance lncRNAs in liquid biopsy samples [25] |
| cDNA Synthesis | High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Provides high-efficiency reverse transcription for challenging samples |
| qPCR Reagents | Power SYBR Green PCR Master Mix | Thermo Fisher Scientific | Enables sensitive detection of lncRNAs with robust amplification |
| Extracellular Vesicle Isolation | Size-exclusion chromatography and ultrafiltration method | Echo Biotech | Isulates EV-associated lncRNAs for cargo analysis [90] |
| Quality Control | Bioanalyzer RNA Integrity Analysis | Agilent Technologies | Assesses RNA quality prior to downstream applications |
| Data Analysis | R/Bioconductor packages (survival, pROC, glmnet) | Open Source | Implements statistical analyses for validation studies [87] [37] |
The selection of appropriate research reagents is critical for successful external validation of lncRNA biomarkers. Specialized RNA isolation kits designed for liquid biopsy samples are essential when working with plasma or serum, as they optimize recovery of low-abundance lncRNAs [25]. High-efficiency cDNA synthesis kits ensure that the limited RNA obtained from clinical samples is adequately converted for subsequent qPCR analysis. For studies focusing on extracellular vesicle-derived lncRNAs, standardized isolation protocols that combine size-exclusion chromatography with ultrafiltration provide reproducible recovery of EV-associated nucleic acids [90]. Computational tools, particularly within the R/Bioconductor environment, offer validated implementations of statistical methods essential for rigorous validation.
Case Studies in External Validation of HCC lncRNA Biomarkers
Case Study 1: PANoptosis-Related lncRNA Signature
A 2025 study developed a PANoptosis-related lncRNA (PRL) prognostic system for HCC and employed a comprehensive external validation strategy. After establishing the signature in the TCGA-LIHC cohort (n=370), researchers validated it in an independent ICGC cohort (n=231), confirming that the high-PRL score group had significantly worse overall survival [87]. The validation included:
- Stratification of ICGC patients into high- and low-risk groups using the same cutoff established in TCGA
- Demonstration of significant survival differences (log-rank p<0.05)
- Multivariate analysis confirming the signature as an independent prognostic factor
- Additional experimental validation through knockdown studies showing suppressed HCC progression
This multi-level validation approach strengthened the evidence for clinical utility of the PRL signature by demonstrating consistent performance across independently generated datasets and providing mechanistic insights through functional studies.
Case Study 2: 4-lncRNA Signature for Early Recurrence Prediction
Another study developed a 4-lncRNA signature (AC108463.1, AF131217.1, CMB9-22P13.1, TMCC1-AS1) for predicting early recurrence in HCC. After construction in the TCGA training set (n=157), the signature was validated in multiple phases [37]:
- Internal validation in the TCGA testing set (n=157)
- External validation in a Jinling Hospital cohort (n=44)
- Functional validation of TMCC1-AS1 in HCC cell lines
The external validation in the clinical cohort confirmed that patients in the high-risk group had significantly higher early recurrence rates than those in the low-risk group. Furthermore, combining the lncRNA signature with established clinical factors (AFP and TNM stage) further improved predictive performance, demonstrating the complementary value of lncRNA biomarkers to existing clinical tools.
External validation through independent cohort studies and public dataset verification represents an indispensable component in the development of clinically useful lncRNA biomarkers for HCC. The integration of computational validation using public repositories with experimental confirmation in well-characterized clinical cohorts provides a robust framework for establishing generalizability and clinical utility. As the field advances, increasing emphasis should be placed on validation across diverse etiologies, stages, and demographic groups to ensure equitable application of lncRNA-based tools. Furthermore, standardization of analytical protocols and reporting standards will enhance comparability across studies and accelerate the translation of promising lncRNA biomarkers from discovery to clinical application.
Hepatocellular carcinoma (HCC) remains a leading cause of cancer-related mortality globally, largely due to limitations in early detection using conventional diagnostic standards. This application note provides a comprehensive comparison between emerging diagnostic approaches integrating machine learning (ML) with long non-coding RNA (lncRNA) biomarkers and traditional methods. We detail experimental protocols for lncRNA quantification and ML model development, present quantitative performance comparisons, and visualize key workflows and molecular pathways. The synthesized evidence demonstrates that ML-driven lncRNA signatures significantly outperform traditional biomarkers like alpha-fetoprotein (AFP) in sensitivity, specificity, and prognostic capability, offering researchers validated methodologies for implementing these advanced diagnostic frameworks in HCC management.
Hepatocellular carcinoma represents a significant global health burden, ranking as the sixth most prevalent cancer worldwide and the fourth most common cause of cancer-related mortality [27]. The disease frequently presents asymptomatically in early stages, often resulting in late diagnosis when treatment options are limited and prognosis is poor [27] [91]. Traditional surveillance protocols rely primarily on abdominal ultrasonography and serum alpha-fetoprotein (AFP) measurement, but these methods face significant limitations including suboptimal sensitivity, operator dependence for ultrasound, and poor performance in specific patient populations such as those with obesity or metabolic dysfunction-associated steatotic liver disease (MASLD) [91].
The emergence of liquid biopsy approaches utilizing circulating biomarkers has opened new avenues for non-invasive HCC detection. Among these, long non-coding RNAs (lncRNAs) - RNA molecules exceeding 200 nucleotides with limited protein-coding potential - have demonstrated considerable promise as cancer biomarkers due to their tissue-specific expression, stability in body fluids, and direct involvement in carcinogenesis [11] [92] [81]. When combined with machine learning algorithms, lncRNA signatures can be integrated with clinical parameters to create powerful predictive models that surpass the diagnostic capabilities of conventional approaches.
Performance Comparison: Quantitative Data Synthesis
Diagnostic Performance Metrics
Table 1: Comparative Performance of Diagnostic Approaches for HCC Detection
| Diagnostic Approach | Sensitivity (%) | Specificity (%) | AUC/Other Metrics | Sample Size | Reference |
|---|---|---|---|---|---|
| Traditional AFP Only | 60-65 | 80-85 | ~0.70-0.75 (AUC) | Varies | [27] [91] |
| Individual lncRNAs | 60-83 | 53-67 | Moderate | 82 participants | [27] |
| ML-lncRNA Integration | 100 | 97 | Superior to all individual markers | 82 participants | [27] |
| 4-lncRNA Signature + AFP + TNM | N/A | N/A | Superior early recurrence prediction | 314 patients | [15] |
| CAIPS (7-gene ML Signature) | N/A | N/A | Highest C-index vs. 150 published signatures | 1,110 patients (6 cohorts) | [93] |
Clinical Application Potential
Table 2: Clinical Applications of Different Diagnostic Paradigms
| Parameter | Traditional Standards | ML-lncRNA Models |
|---|---|---|
| Early Detection Capability | Limited (misses >1/3 early cases) | Enhanced (100% sensitivity reported) |
| Prognostic Prediction | Limited to tumor staging | Strong early recurrence prediction |
| Therapeutic Guidance | Limited | Predicts response to TACE, targeted therapy, immunotherapy |
| Implementation Barriers | Low cost, widespread availability | Requires specialized computational resources |
| Biomarker Stability | Moderate | High (lncRNAs stable in circulation) |
Experimental Protocols and Methodologies
Protocol 1: Plasma lncRNA Quantification and Analysis
Principle: Circulating lncRNAs can be reliably isolated from plasma samples and quantified using qRT-PCR, providing measurable biomarkers for HCC detection and monitoring.
Materials and Reagents:
- miRNeasy Mini Kit (QIAGEN, cat no. 217004)
- RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, cat no. K1622)
- PowerTrack SYBR Green Master Mix (Applied Biosystems, cat no. A46012)
- Primers for target lncRNAs (LINC00152, LINC00853, UCA1, GAS5)
- GAPDH primers for normalization
- ViiA 7 real-time PCR system or equivalent
Procedure:
- Sample Collection and Processing: Collect peripheral blood in EDTA-containing tubes. Process within 2 hours of collection by centrifugation at 2,000 × g for 10 minutes at 4°C. Transfer plasma to clean tubes and centrifuge at 12,000 × g for 10 minutes to remove cellular debris. Store at -80°C until RNA extraction.
- RNA Isolation: Use miRNeasy Mini Kit according to manufacturer's protocol. Include DNase treatment to eliminate genomic DNA contamination. Elute RNA in 30-50 μL RNase-free water.
- cDNA Synthesis: Perform reverse transcription using RevertAid First Strand cDNA Synthesis Kit with 1μg total RNA input in 20μL reaction volume.
- qRT-PCR Analysis: Prepare reactions in triplicate using PowerTrack SYBR Green Master Mix. Use the following cycling conditions: 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Include no-template controls for each primer set.
- Data Analysis: Calculate relative expression using the 2−ΔΔCT method with GAPDH as endogenous control [27].
Technical Notes:
- Maintain consistent sample processing times to minimize pre-analytical variability.
- Include inter-plate calibrators for experiments run across multiple plates.
- Establish reproducibility with coefficient of variation <10% for replicate samples.
- Consider using spike-in controls for RNA isolation efficiency monitoring.
Protocol 2: Machine Learning Model Development for HCC Diagnosis
Principle: Integration of lncRNA expression data with clinical parameters using machine learning algorithms enhances diagnostic and prognostic accuracy for HCC.
Materials and Software:
- Python with Scikit-learn library
- Clinical dataset including lncRNA expression, liver function tests, and patient outcomes
- High-performance computing environment for large-scale analysis
Procedure:
- Data Preprocessing:
- Compile dataset with lncRNA expression values (LINC00152, LINC00853, UCA1, GAS5) and clinical parameters (ALT, AST, AFP, bilirubin, albumin)
- Perform data normalization using z-scores or quantile normalization
- Handle missing values using appropriate imputation methods
- Split data into training (70%) and validation (30%) sets
Feature Selection:
- Apply multiple machine learning algorithms including Least Absolute Shrinkage and Selection Operator (LASSO), Random Forest, and Support Vector Machine Recursive Feature Elimination (SVM-RFE)
- Identify most predictive features using cross-validation
- Reduce dimensionality while maintaining predictive power
Model Construction:
- Develop ensemble models combining multiple algorithms
- Optimize hyperparameters using grid search with cross-validation
- Validate model performance on independent validation set
- Assess feature importance using SHAP analysis [93]
Performance Validation:
- Evaluate using receiver operating characteristic (ROC) analysis
- Calculate sensitivity, specificity, positive and negative predictive values
- Compare with traditional diagnostic methods using DeLong's test
- Perform external validation in independent cohorts when possible
Technical Notes:
- Address class imbalance using SMOTE or similar techniques if needed
- Implement rigorous cross-validation to prevent overfitting
- Consider time-dependent ROC analysis for prognostic models
- Utilize multi-center cohorts to enhance generalizability
Diagram Title: ML-lncRNA Model Development Workflow
Molecular Mechanisms and Pathway Visualization
LncRNAs contribute to hepatocarcinogenesis through diverse molecular mechanisms, functioning as both oncogenic drivers and tumor suppressors. Key oncogenic lncRNAs include HULC, HOTAIR, MALAT1, and UCA1, while tumor-suppressive lncRNAs include GAS5 and others [11] [94]. These molecules regulate critical cellular processes through multiple mechanisms:
4.1 Epigenetic Regulation: LncRNAs such as HOTAIR interact with Polycomb Repressive Complex 2 (PRC2) to mediate histone H3 lysine-27 trimethylation, leading to transcriptional repression of tumor suppressor genes [11] [81].
4.2 miRNA Sponging: LncRNAs including HULC function as competitive endogenous RNAs (ceRNAs) that sequester microRNAs, preventing them from binding to their target mRNAs. HULC specifically downregulates miR-372 and miR-186, thereby modulating expression of their target genes [94].
4.3 Protein Interactions: LncRNAs can serve as scaffolds that bring multiple proteins together to form functional complexes. For example, the lncRNA ANRIL forms complexes with chromatin-modifying proteins that regulate the INK4/ARF tumor suppressor locus [94].
4.4 Autophagy Regulation: Multiple lncRNAs modulate autophagic flux in HCC through pathways including PI3K/AKT/mTOR, AMPK, and Beclin-1. This regulation contributes to the dual role of autophagy in HCC - acting as a tumor suppressor in early stages but promoting survival in advanced disease [95].
Diagram Title: LncRNA Mechanisms in HCC Pathogenesis
Table 3: Key Research Reagents and Resources for ML-lncRNA HCC Studies
| Category | Specific Product/Kit | Application Purpose | Technical Notes |
|---|---|---|---|
| RNA Isolation | miRNeasy Mini Kit (QIAGEN) | Total RNA extraction from plasma/serum | Includes DNase treatment; suitable for low-abundance RNAs |
| cDNA Synthesis | RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) | Reverse transcription for qRT-PCR | Use random hexamers for lncRNA detection |
| qRT-PCR Master Mix | PowerTrack SYBR Green Master Mix (Applied Biosystems) | Quantitative lncRNA expression analysis | Optimized for difficult templates |
| PCR Platform | ViiA 7 Real-Time PCR System (Applied Biosystems) | High-throughput lncRNA quantification | Alternative: CFX96 (Bio-Rad) |
| Machine Learning | Python Scikit-learn Library | ML model development and validation | Open-source; comprehensive algorithm collection |
| Statistical Analysis | R with survival, pROC packages | Statistical analysis and visualization | Essential for survival and ROC analyses |
The integration of machine learning with lncRNA biomarker profiles represents a paradigm shift in HCC diagnosis that substantially outperforms traditional diagnostic standards. The documented performance metrics demonstrate clear advantages in sensitivity, specificity, and prognostic capability, with ML-lncRNA models achieving up to 100% sensitivity and 97% specificity compared to 60-65% sensitivity for AFP alone [27]. These approaches leverage the biological relevance and stability of lncRNAs in circulation while harnessing the pattern recognition power of machine learning algorithms.
Future developments in this field will likely focus on several key areas: (1) validation of multi-lncRNA signatures in large, diverse patient cohorts to establish clinical utility across different etiologies and ethnicities; (2) integration of multi-omics data including genomic, proteomic, and metabolomic markers to further enhance diagnostic accuracy; (3) development of point-of-care testing platforms to enable widespread clinical implementation; and (4) exploration of lncRNAs as therapeutic targets in addition to diagnostic markers.
For researchers implementing these approaches, we recommend rigorous adherence to standardized protocols for pre-analytical sample processing, utilization of multiple validation cohorts, and transparent reporting of ML model architectures and performance metrics. As these technologies continue to mature, ML-lncRNA integration holds significant promise for transforming HCC management through earlier detection, accurate prognosis prediction, and ultimately improved patient outcomes.
Hepatocellular carcinoma (HCC) represents a significant global health challenge, ranking as the sixth most commonly diagnosed cancer and the fourth leading cause of cancer-related mortality worldwide [37] [96]. A critical factor impacting survival outcomes is cancer recurrence, with approximately 70% of patients experiencing recurrence within five years of surgical resection [37] [96]. Clinically, recurrence within two years post-surgery is classified as early recurrence, which carries a significantly poorer prognosis compared to late recurrence [37]. This distinction makes the prediction of early recurrence a crucial focus for improving clinical management and survival outcomes.
Long non-coding RNAs (lncRNAs) have emerged as promising molecular biomarkers for cancer prognosis. These RNA transcripts, exceeding 200 nucleotides in length without protein-coding capacity, are intensively involved in HCC progression through diverse mechanisms including binding with RNA, DNA, proteins, or encoding small peptides [37]. Their differential expression patterns in cancer tissues and stability in circulating biofluids make them particularly suitable for diagnostic and prognostic applications [6] [25]. The integration of lncRNA profiling with machine learning algorithms represents a transformative approach for developing robust predictive models that can stratify patients according to recurrence risk, potentially enabling more personalized treatment strategies and enhanced post-surgical surveillance [37] [6].
Multiple research groups have developed and validated multi-lncRNA signatures for predicting early recurrence in HCC. The table below summarizes key prognostic signatures reported in recent literature:
Table 1: Validated lncRNA Signatures for HCC Early Recurrence Prediction
| Signature Size | Specific lncRNAs | AUC/Performance | Clinical Utility | Reference |
|---|---|---|---|---|
| 4-lncRNA | AC108463.1, AF131217.1, CMB9-22P13.1, TMCC1-AS1 | Combination with AFP and TNM improved predictive performance | Excellent predictability when combined with standard clinical markers | [37] |
| 25-lncRNA | Not fully specified | Superior to individual clinical factors | Best predictive performance among individual risk factors; synergizes with AFP, TNM, and vascular invasion | [96] |
| 9-IR-lncRNA | Immune-related lncRNAs | Validated in testing cohort | Important clinical implications for individualized treatment guidance | [97] |
| Panel of 4 | LINC00152, LINC00853, UCA1, GAS5 | 100% sensitivity, 97% specificity in ML model | Machine learning integration with conventional biomarkers for diagnosis | [6] |
Meta-analytical data further substantiates the prognostic value of lncRNAs in HCC, demonstrating that patients with elevated expression of oncogenic lncRNAs experience significantly poorer overall survival (pooled HR: 1.25) and recurrence-free survival (pooled HR: 1.66) [98]. The consistency of these findings across multiple study designs highlights the robustness of lncRNAs as prognostic biomarkers.
Experimental Protocols for lncRNA Biomarker Development
Computational Identification of Recurrence-Associated lncRNAs
The development of a prognostic lncRNA signature begins with comprehensive bioinformatic analysis using RNA sequencing data from cohorts of HCC patients with complete clinical follow-up information.
Table 2: Key Computational Methods for lncRNA Signature Development
| Method | Purpose | Key Parameters | Implementation |
|---|---|---|---|
| Differential Expression | Identify lncRNAs differentially expressed between tumor and normal tissues | |log2FC| > 1, FDR < 0.05 | DESeq2, edgeR, or limma R packages |
| Survival Analysis | Select lncRNAs associated with recurrence-free survival | P < 0.05 | Univariate Cox regression via "survival" R package |
| Machine Learning Feature Selection | Reduce dimensionality and select most predictive lncRNAs | Lambda.min for LASSO; 5-fold cross-validation for SVM-RFE; top features for random forest | LASSO, random forest, and SVM-RFE algorithms |
| Multivariate Cox Regression | Finalize signature and calculate coefficients | P < 0.05 | "survival" R package to establish risk score formula |
The standard risk score calculation formula is: Risk Score = Σ (lncRNA expression × corresponding coefficient). Patients are then stratified into high-risk and low-risk groups using the median risk score as the cutoff threshold [37] [96]. Model performance is evaluated using time-dependent receiver operating characteristic (ROC) curves and Kaplan-Meier survival analysis with log-rank tests to assess the significance of survival differences between risk groups [37].
Wet-Lab Validation Protocol
Following computational identification, candidate lncRNAs require validation using clinically applicable methods:
Sample Collection and RNA Extraction
- Collect plasma samples from HCC patients and age-matched healthy controls (500 μL per sample) [6] [25]
- Extract total RNA using commercial kits (e.g., miRNeasy Mini Kit or Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit)
- Treat RNA samples with DNase to remove genomic DNA contamination
- Assess RNA quality and quantity using spectrophotometry
cDNA Synthesis and Quantitative RT-PCR
- Perform reverse transcription using High-Capacity cDNA Reverse Transcription Kit
- Conduct quantitative real-time PCR with Power SYBR Green PCR Master Mix
- Use the following cycling conditions: initial denaturation at 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 62°C for 1 minute [25]
- Include housekeeping genes (GAPDH or β-actin) for normalization
- Analyze each sample in triplicate with appropriate no-template controls
- Calculate relative expression using the 2−ΔΔCt method [6] [25]
Analytical Validation
- Perform ROC curve analysis to evaluate diagnostic accuracy of individual lncRNAs
- Assess sensitivity and specificity at optimal cutoff values
- For machine learning integration: combine lncRNA expression data with clinical parameters (AFP, ALT, AST, bilirubin, albumin) using algorithms such as XGBoost [6] [99]
- Validate predictive models in independent patient cohorts to ensure generalizability
Visualizing Experimental Workflows
The following diagrams illustrate key procedural workflows and molecular relationships in lncRNA biomarker development:
Figure 1: Comprehensive Workflow for lncRNA Signature Development
Figure 2: Molecular Pathways to HCC Recurrence
Research Reagent Solutions
Table 3: Essential Research Reagents for lncRNA Biomarker Studies
| Reagent Category | Specific Product Examples | Application Purpose | Key Considerations |
|---|---|---|---|
| RNA Extraction Kits | miRNeasy Mini Kit (QIAGEN), Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit (Norgen Biotek) | Isolation of high-quality total RNA from tissues or plasma | Preserve RNA integrity; effectively recover small RNAs |
| Reverse Transcription Kits | High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher), RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) | Generate cDNA for downstream qPCR applications | Ensure efficient transcription of long RNA species |
| qPCR Master Mixes | Power SYBR Green PCR Master Mix (Thermo Fisher), PowerTrack SYBR Green Master Mix (Applied Biosystems) | Quantitative detection of lncRNA expression | Provide high sensitivity and specificity |
| Reference Genes | GAPDH, β-actin | Normalization of lncRNA expression data | Validate stability in specific sample matrices |
| Primer Sets | Custom-designed lncRNA-specific primers | Target amplification in qPCR assays | Verify specificity for intended lncRNA transcripts |
The integration of lncRNA biomarkers with machine learning algorithms represents a paradigm shift in prognostic assessment for hepatocellular carcinoma. The protocols outlined herein provide a standardized framework for developing and validating lncRNA-based predictive models that can stratify HCC patients according to their risk of early recurrence. These approaches demonstrate superior performance compared to conventional clinical markers alone, offering the potential for more personalized postoperative management, including tailored surveillance protocols and adjuvant therapy selection for high-risk patients.
Future directions in this field should focus on the standardization of analytical protocols across institutions, the development of point-of-care detection platforms, and the integration of lncRNA signatures with other molecular biomarker classes to create comprehensive prognostic models. As validation studies continue to accumulate, lncRNA-based prognostic tools are poised to become invaluable clinical assets in the ongoing effort to improve survival outcomes for HCC patients.
Hepatocellular carcinoma (HCC) represents a significant global health challenge, ranking as the sixth most common cancer worldwide and the fourth leading cause of cancer-related mortality [6]. The current diagnostic landscape relies heavily on imaging techniques like ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI), supplemented by the serum biomarker alpha-fetoprotein (AFP). However, these methods present considerable limitations for early detection, with ultrasound sensitivity as low as 50% for early lesions and small tumor nodules [54]. This diagnostic challenge creates a critical need for more precise, non-invasive biomarkers that can detect HCC at curative stages.
Long non-coding RNAs (lncRNAs) have emerged as promising biomarker candidates, with studies demonstrating their differential expression patterns across diverse cancers, affecting tumor growth and survival potential [6]. The integration of machine learning (ML) approaches for analyzing these molecular signatures offers a transformative pathway toward developing robust diagnostic tools. This document outlines a comprehensive framework for advancing lncRNA-based ML models through regulatory milestones toward clinical implementation, providing researchers with validated protocols and assessment criteria.
Performance Benchmarks: Current and Emerging Biomarkers
Table 1: Comparative Performance of HCC Diagnostic Modalities
| Diagnostic Method | Sensitivity Range | Specificity Range | Key Advantages | Notable Limitations |
|---|---|---|---|---|
| Ultrasound | ~50% (early lesions) | >90% | Non-invasive, widely available | Limited sensitivity for small tumors [54] |
| CT/MRI | >90% (tumors >2cm) | >90% | High accuracy for established tumors | High cost, not suitable for routine screening [54] |
| AFP Serology | 60-80% | 80-90% | Low cost, standardized | Elevated in benign liver conditions [6] [54] |
| Individual lncRNAs (LINC00152, UCA1, etc.) | 60-83% | 53-67% | Cancer-specific, detectable in plasma | Moderate individual performance [6] |
| ML-Integrated Panels (lncRNAs + clinical variables) | Up to 100% | Up to 97% | Multi-analyte approach, high accuracy | Computational complexity, requires validation [6] [14] |
Table 2: Experimental Performance of ML Models in HCC Diagnosis
| Machine Learning Model | Reported Accuracy | Sample Size (Training/Testing) | Key Features Integrated |
|---|---|---|---|
| Logistic Regression | 92% AUC | 287/72 (external validation) | Clinical factors + metabolites [100] |
| Light Gradient Boosting Machine (LGBM) | 98.75% | 187/80 | RNA signatures + clinical data [14] |
| Random Forest | 96.25% | 187/80 | RNA signatures + clinical data [14] |
| Python Scikit-learn Platform | 100% sensitivity, 97% specificity | 52 HCC patients, 30 controls | 4 lncRNAs + clinical laboratory parameters [6] |
| Deep Neural Networks (DNN) | 91.25% | 187/80 | RNA signatures + clinical data [14] |
Regulatory Framework and Readiness Criteria
Navigating the regulatory pathway requires meticulous planning and adherence to quality standards from discovery through clinical implementation. The FDA's Chemistry, Manufacturing, and Controls Development and Readiness Pilot (CDRP) Program provides a valuable framework for expedited development, emphasizing increased communication between sponsors and regulatory agencies [101]. For diagnostic applications, readiness encompasses both analytical and clinical validation, with increasing evidence requirements through each development phase.
Foundational Regulatory Principles
The core principle of regulatory readiness involves embedding compliance into daily operations rather than treating it as a last-minute preparation [102] [103]. Documentation must tell a coherent quality and compliance story, with every batch record, deviation, and Corrective and Preventive Action (CAPA) clearly demonstrating decision-making processes and their connection to patient safety and product quality [102]. Personnel competency is equally crucial, with team members able to articulate their roles, explain decisions, and demonstrate understanding of quality principles beyond mere procedure memorization [102].
Clinical Trial Compliance Framework
For biomarkers intended to support therapeutic development, clinical trial compliance requires attention to several interdependent domains. Regulatory documentation must remain current, complete, and readily accessible, with particular emphasis on informed consent procedures, protocol adherence, and safety reporting [103]. Best practices include conducting internal audits and mock inspections, adopting standardized document management systems, and maintaining strict version control [103]. Common inspection findings include missing or incomplete signatures, insufficient delegation documentation, and delays in safety reporting, all of which should be addressed proactively [103].
Experimental Protocols for lncRNA Biomarker Development
Sample Collection and RNA Extraction Protocol
Principle: Obtain high-quality plasma samples and extract total RNA while preserving lncRNA integrity for downstream applications.
Materials:
- EDTA or sodium citrate blood collection tubes
- Centrifuge capable of 4°C operation
- miRNeasy Mini Kit (Qiagen, cat no. 217004) or equivalent
- Polypropylene tubes for plasma storage
- -80°C freezer for sample preservation
Procedure:
- Collect venous blood into sodium citrate tubes and centrifuge at 4°C, 3000 rpm for 20 minutes within one hour of collection [14].
- Aliquot supernatant plasma into polypropylene tubes without disturbing the buffy coat.
- Store plasma at -80°C until RNA extraction.
- Extract total RNA using miRNeasy Mini Kit according to manufacturer's protocol [6] [14].
- Validate RNA quality and purity using Qubit 3.0 Fluorimeter with appropriate assay kits [14].
Technical Notes:
- Consistent processing time is critical to prevent RNA degradation.
- For the Biocrates Absolute IDQ p180 kit, follow established protocols for metabolite quantification parallel to RNA analysis [100].
- Document all sample handling procedures to meet quality standards for regulatory submissions [102].
cDNA Synthesis and Quantitative Real-Time PCR
Principle: Convert extracted RNA to cDNA and quantify lncRNA expression levels using specific primers.
Materials:
- RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, cat no. K1622)
- T100 thermal cycler (Bio-Rad) or equivalent
- PowerTrack SYBR Green Master Mix (Applied Biosystems, cat no. A46012)
- ViiA 7 real-time PCR system (Applied Biosystems) or equivalent
- Sequence-specific primers for target lncRNAs (LINC00152, LINC00853, UCA1, GAS5)
Procedure:
- Perform reverse transcription using 500ng total RNA and RevertAid First Strand cDNA Synthesis Kit on a thermal cycler [6].
- Prepare qRT-PCR reactions in triplicate using PowerTrack SYBR Green Master Mix according to manufacturer specifications.
- Run reactions on ViiA 7 real-time PCR system with the following cycling conditions:
- Initial denaturation: 95°C for 10 minutes
- 40 cycles of: 95°C for 15 seconds, 60°C for 60 seconds
- Use GAPDH as the endogenous control for normalization [6].
- Calculate relative expression using the 2^(-ΔΔCt) method [14].
Technical Notes:
- Include no-template controls to detect contamination.
- Establish standard curves for efficiency calculations.
- Document all protocol deviations for regulatory compliance [103].
Machine Learning Model Development Protocol
Principle: Develop and validate a predictive model integrating lncRNA expression data with clinical variables.
Materials:
- Python programming environment with scikit-learn, Pandas, NumPy libraries
- Clinical and lncRNA expression dataset with appropriate sample size
- Computational resources sufficient for model training (multi-core processors, adequate RAM)
Procedure:
- Data Preprocessing:
Dataset Partitioning:
- Allocate 80% of data for model training/validation (using tenfold cross-validation)
- Reserve 20% for external validation testing [100].
Model Training and Evaluation:
- Implement multiple algorithms (Logistic Regression, Random Forest, SVM, XGBoost, etc.)
- Optimize hyperparameters through cross-validation
- Evaluate models using accuracy, sensitivity, specificity, and AUC metrics
- Compare performance against clinical-only models to assess added value [104]
Technical Notes:
- Apply recursive feature elimination with cross-validation to identify optimal feature sets [100].
- Ensure data security and privacy compliance throughout analysis [104].
- Document all modeling decisions and parameter settings for regulatory review [102].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagents and Platforms for lncRNA Biomarker Development
| Reagent/Platform | Manufacturer | Function | Application Notes |
|---|---|---|---|
| miRNeasy Mini Kit | Qiagen | Total RNA isolation from plasma/serum | Preserves lncRNA integrity; compatible with small volumes [6] [14] |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific | Reverse transcription | Efficient conversion of lncRNAs to cDNA [6] |
| PowerTrack SYBR Green Master Mix | Applied Biosystems | qRT-PCR detection | Sensitive detection of lncRNA amplification [6] |
| Absolute IDQ p180 Kit | Biocrates | Targeted metabolite quantification | Enables multi-omics integration with lncRNA data [100] |
| ViiA 7 Real-Time PCR System | Applied Biosystems | High-throughput qPCR | 384-well format for large-scale validation studies [6] |
| Python Scikit-learn | Open Source | Machine learning implementation | Comprehensive algorithms for predictive model development [6] [100] |
| Qubit 3.0 Fluorimeter | Invitrogen | Nucleic acid quantification | Accurate RNA concentration measurements [14] |
Pathway to Clinical Implementation
The clinical implementation pathway requires systematic progression through validation milestones. The initial discovery phase should prioritize lncRNAs with strong biological rationale, such as those involved in autophagy regulation or disulfidptosis, a newly discovered form of programmed cell death [105] [95]. Analytical validation must establish assay precision, accuracy, sensitivity, and specificity under controlled conditions, while clinical validation demonstrates performance in intended-use populations.
Engaging with regulatory agencies through mechanisms like the CDRP Program facilitates early alignment on development strategies and validation requirements [101]. Pivotal studies should be designed with input from both regulators and clinical stakeholders to ensure endpoints address real-world diagnostic needs. Following regulatory approval, implementation requires integration into clinical workflows, establishment of reimbursement pathways, and education of healthcare providers on appropriate use contexts.
The integration of machine learning with lncRNA biomarkers represents a promising frontier in HCC diagnostics, with demonstrated potential to exceed the performance of current standard approaches. Successful clinical implementation requires not only technical excellence but also rigorous adherence to regulatory pathways and quality standards. By following the structured framework presented in this document, researchers can systematically address both scientific and regulatory requirements, accelerating the translation of promising biomarkers from discovery to clinical practice where they can impact patient outcomes through earlier and more accurate HCC detection.
Conclusion
The integration of machine learning with lncRNA biomarkers represents a paradigm shift in hepatocellular carcinoma diagnostics, demonstrating unprecedented accuracy that far surpasses traditional methods like AFP. The synthesis of evidence reveals that ML-driven models can achieve remarkable diagnostic performance, with studies reporting sensitivities up to 100% and specificities of 97-98.75% by effectively analyzing complex lncRNA expression patterns. Future directions must focus on multi-center prospective validations in diverse patient populations, standardization of liquid biopsy protocols, and the development of reproducible, interpretable AI models that clinicians can trust. The successful translation of these technologies from research to clinical practice holds immense potential to revolutionize early HCC detection, enable personalized treatment strategies based on molecular subtyping, and ultimately significantly improve survival rates for this deadly cancer. Researchers and drug developers should prioritize creating unified data standards and collaborative frameworks to accelerate this promising field toward clinical implementation.
References
- 1. Long non-coding RNAs: definitions, functions, challenges ... [https://www.nature.com/articles/s41580-022-00566-8]
- 2. Exploring long noncoding RNAs: structure, function ... [https://www.revvity.com/blog/exploring-long-noncoding-rnas-structure-function-biomarker-potential-and-sequencing]
- 3. Long Non-Coding RNAs: Tools for Understanding and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9564174/]
- 4. Gene regulation by long non-coding RNAs and its biological ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7754182/]
- 5. Noncoding RNA - Applications [https://horizondiscovery.com/en/applications/rnai/noncoding-rna-applications]
- 6. Analysis of four long non-coding RNAs for hepatocellular ... [https://www.nature.com/articles/s41598-024-80926-w]
- 7. Gene regulation by long non-coding RNAs and its ... [https://www.nature.com/articles/s41580-020-00315-9]
- 8. Machine learning-based identification of diagnostic and ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0331118]
- 9. Integrating AI and RNA biomarkers in cancer [https://pmc.ncbi.nlm.nih.gov/articles/PMC12512311/]
- 10. The epigenetic regulatory network of long noncoding RNAs ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12205805/]
- 11. Long non-coding RNA in hepatocellular carcinoma [https://link.springer.com/article/10.1007/s13402-025-01119-9]
- 12. Long non-coding RNAs and autophagy: dual drivers of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12339970/]
- 13. Deciphering the nexus between long non-coding RNAs ... [https://www.nature.com/articles/s41420-024-02200-2]
- 14. Machine-Learning-Based Identification of Key Feature ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11268357/]
- 15. Combining a machine-learning derived 4-lncRNA signature ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-023-09194-8]
- 16. The role of long non-coding RNA in hepatocellular carcinoma [https://pmc.ncbi.nlm.nih.gov/articles/PMC10929815/]
- 17. LncRNAs expression signatures of hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4033470/]
- 18. promotes the growth of lncRNA ... HULC hepatocellular carcinoma [https://pubmed.ncbi.nlm.nih.gov/28634076/]
- 19. HBx-upregulated lncRNA UCA1 promotes cell growth and ... [https://www.nature.com/articles/srep23521]
- 20. Long noncoding RNA MALAT1 promotes hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5334181/]
- 21. Role of the long non-coding RNA HOTAIR in hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5529952/]
- 22. LINC00152 promotes proliferation in hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4767473/]
- 23. LncRNA GAS5 enhances radiosensitivity of hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8506991/]
- 24. Proceedings of cell-free noncoding RNA biomarker studies in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12495990/]
- 25. lncRNAs as Biomarkers of Hepatocellular Carcinoma Risk ... [https://www.mdpi.com/1467-3045/47/5/348]
- 26. Liquid biopsy in cancer: current status, challenges and ... [https://www.nature.com/articles/s41392-024-02021-w]
- 27. Analysis of four long non-coding RNAs for hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11604705/]
- 28. A LncRNA panel within EpCAM-specific exosomes for ... [https://respiratory-research.biomedcentral.com/articles/10.1186/s12931-025-03220-x]
- 29. Plasma exosomal lncRNA-related signatures define ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12568545/]
- 30. Machine learning-based disulfidptosis-related lncRNA ... [https://www.nature.com/articles/s41598-024-54115-8]
- 31. Circulating blood biomarkers for minimal residual disease ... [https://www.sciencedirect.com/science/article/pii/S0305737225000301]
- 32. Development of a novel routine blood-based AI model for ... [https://www.sciencedirect.com/science/article/pii/S2949819825001104]
- 33. lncRNAs as Biomarkers of Hepatocellular Carcinoma Risk ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12110020/]
- 34. Machine learning-based hepatocellular carcinoma risk ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12504168/]
- 35. Accessing and Downloading TCGA Data - BTEP Coding Club [https://bioinformatics.ccr.cancer.gov/docs/btep-coding-club/CC2024/TCGA/TCGA_download/]
- 36. How To Download Gene Expression Data From TCGA and ... [https://ngs101.com/how-to-download-gene-expression-data-from-tcga-and-gtex-using-r-a-complete-guide-for-cancer-genomics-research/]
- 37. Combining a machine-learning derived 4-lncRNA signature ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9972730/]
- 38. Identification of diagnostic long non‑coding RNA ... [https://www.spandidos-publications.com/10.3892/mmr.2019.10307]
- 39. Identification of lncRNA biomarkers for lung cancer through ... [https://www.aging-us.com/article/103496/text]
- 40. Diagnosis, prognosis and bioinformatics analysis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5707062/]
- 41. Machine-Learning-Based Identification of Key Feature ... [https://www.sciencedirect.com/science/article/abs/pii/S0973688324001130]
- 42. TANRIC [https://bioinformatics.mdanderson.org/public-software/tanric/]
- 43. Identification and characterization of a 25-lncRNA prognostic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8556945/]
- 44. Comprehensive analysis of basement membrane and ... [https://www.sciencedirect.com/science/article/pii/S2405844023076703]
- 45. LightGBM Explained: Faster and Smarter Gradient Boosting [https://medium.com/@abhaysingh71711/lightgbm-explained-faster-and-smarter-gradient-boosting-4ab549629925]
- 46. LightGBM: A Guide [https://builtin.com/articles/lightgbm]
- 47. Support Vector Machine (SVM) Algorithm [https://www.geeksforgeeks.org/machine-learning/support-vector-machine-algorithm/]
- 48. 10 Pros & Cons of Support Vector Machines [2025] [https://digitaldefynd.com/IQ/pros-cons-of-support-vector-machines/]
- 49. Algorithm in Machine Learning - GeeksforGeeks Random Forest [https://www.geeksforgeeks.org/machine-learning/random-forest-algorithm-in-machine-learning/]
- 50. What Are the Advantages and Disadvantages of Random ? Forest [https://www.coursera.org/articles/advantages-and-disadvantages-of-random-forest]
- 51. What are Neural Networks? — updated 2025 | IxDF [https://www.interaction-design.org/literature/topics/neural-networks?srsltid=AfmBOopqP5XSLRHCqNbWDatQY0naRH7gDY-EMvR2HILai0TXgisdBknr]
- 52. What is a Neural Network? [https://www.geeksforgeeks.org/machine-learning/neural-networks-a-beginners-guide/]
- 53. Minimal clinical predictors enable machine learning ... [https://www.nature.com/articles/s41598-025-22927-x]
- 54. Biomarkers for diagnosis and therapeutic options in ... [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-024-02101-z]
- 55. Machine Learning Identifies a Signature of Nine Exosomal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10377993/]
- 56. Long Non-Coding RNAs: Potential Biomarkers and Targets ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7757045/]
- 57. Potential lncRNA Biomarkers for HBV-Related ... [https://pubmed.ncbi.nlm.nih.gov/34692847/]
- 58. Identification of a novel prognostic signature for HCC and ... [https://www.nature.com/articles/s41598-022-13792-z]
- 59. Four Autophagy-Related lncRNAs Predict the Prognosis of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7568797/]
- 60. Identification of prognostic long non-coding RNA signature ... [https://www.aging-us.com/article/203322/text]
- 61. Characteristics of extracellular vesicle-derived lncRNAs ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12619285/]
- 62. The prognostic correlation of AFP level at diagnosis with ... [https://www.nature.com/articles/s41598-017-12834-1]
- 63. LncRNACNVIntegrateR: a novel framework for correlating ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12535741/]
- 64. Prediction of survival and analysis of prognostic factors for ... [https://bmcgastroenterol.biomedcentral.com/articles/10.1186/s12876-024-03185-z]
- 65. Competing Risk Bias in Prognostic Models Predicting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11307513/]
- 66. Feature reduction for hepatocellular carcinoma prediction ... [https://journalofbigdata.springeropen.com/articles/10.1186/s40537-024-00944-3]
- 67. A Machine Learning Model to Predict Risk for ... [https://www.sciencedirect.com/science/article/pii/S2772572324000074]
- 68. AI enhanced diagnostic accuracy and workload reduction ... [https://www.nature.com/articles/s41746-025-01892-9]
- 69. A Phase 3 Biomarker Validation of GALAD for the Detection of ... [https://pubmed.ncbi.nlm.nih.gov/39293548/]
- 70. Diagnostic Value of lncRNAs as Biomarker in Hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6205093/]
- 71. Identifying novel transcript biomarkers for hepatocellular carcinoma (HCC) using RNA-Seq datasets and machine learning | BMC Cancer | Full Text [https://bmccancer.biomedcentral.com/articles/10.1186/s12885-021-08704-9]
- 72. Identifying novel transcript biomarkers for hepatocellular carcinoma (HCC) using RNA-Seq datasets and machine learning - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8394105/]
- 73. Long non-coding RNAs as a biomarker for homologous ... [https://www.nature.com/articles/s42003-025-08836-9]
- 74. Discovering novel prognostic biomarkers of hepatocellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11737334/]
- 75. Review Artificial Intelligence in Hepatology [https://www.sciencedirect.com/science/article/pii/S2772947825000635]
- 76. Long non-coding RNA-based single and combination ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12405140/]
- 77. Development and validation of an interpretable machine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11899164/]
- 78. AI-driven multi-omics integration in precision oncology [https://pmc.ncbi.nlm.nih.gov/articles/PMC12634751/]
- 79. Long non-coding RNA-based single and combination ... [https://link.springer.com/article/10.1007/s12672-025-03309-1]
- 80. Screening and validation of long non-coding RNAs ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12214067/]
- 81. Blood-derived lncRNAs as biomarkers for cancer diagnosis [https://www.nature.com/articles/s41698-022-00283-7]
- 82. Development and validation of a small-sample machine ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12211751/]
- 83. An Artificial Intelligence Pipeline for Hepatocellular Carcinoma [https://pmc.ncbi.nlm.nih.gov/articles/PMC12229156/]
- 84. Long nonâ€coding RNA as a potential diagnostic and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10844705/]
- 85. A novel disulfidptosis-related lncRNA signature for ... [https://pubmed.ncbi.nlm.nih.gov/41261580/]
- 86. Investigating diagnostic potential of long non-coding RNAs ... [https://www.nature.com/articles/s41598-024-57987-y]
- 87. Identification and validation of PANoptosis-related LncRNAs prognosis... [https://www.nature.com/articles/s41598-025-90498-y?error=cookies_not_supported&code=96fdb91b-ce85-4272-a50a-a529a404c31c]
- 88. Amino acid metabolism-related LncRNAs as prognostic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12545943/]
- 89. Integrating bioinformatics and experimental validation to ... [https://cancerci.biomedcentral.com/articles/10.1186/s12935-023-03208-x]
- 90. Characteristics of extracellular vesicle-derived lncRNAs during the... [https://bmccancer.biomedcentral.com/articles/10.1186/s12885-025-15237-y]
- 91. Novel Biomarkers for Early Detection of Hepatocellular ... [https://www.mdpi.com/2075-4418/14/20/2278]
- 92. Unveiling the Role of LncRNAs in Tumorigenesis [https://www.sciencedirect.com/science/article/pii/S305078712500085X]
- 93. a multi-center and large-scale study | npj Precision Oncology [https://www.nature.com/articles/s41698-025-01010-8]
- 94. A Systematic Review of Long Noncoding RNAs in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6076971/]
- 95. Long non-coding RNAs and autophagy: dual drivers of ... [https://www.nature.com/articles/s41420-025-02667-7]
- 96. Identification and characterization of a 25-lncRNA prognostic ... [https://bmccancer.biomedcentral.com/articles/10.1186/s12885-021-08827-z]
- 97. Construction of an HCC recurrence model based on the ... [https://www.sciencedirect.com/science/article/pii/S216225312100281X]
- 98. lncRNAs as prognostic molecular biomarkers in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5601763/]
- 99. Prediction of long-term recurrence-free and overall survival ... [https://www.nature.com/articles/s41698-025-00978-7]
- 100. Machine learning approaches for biomarker discovery to ... [https://www.nature.com/articles/s41598-023-42338-0]
- 101. Chemistry, Manufacturing, and Controls Development ... [https://www.fda.gov/drugs/pharmaceutical-quality-resources/chemistry-manufacturing-and-controls-development-and-readiness-pilot-cdrp-program]
- 102. FDA Inspection Readiness in 2025: A Complete Guide & ... [https://www.thefdagroup.com/blog/how-to-prepare-for-an-fda-inspection]
- 103. Clinical Trial Compliance: Preparing for FDA Inspection ... [https://www.srainternational.org/blogs/srai-news/2025/10/09/clinical-trial-compliance-fda-inspection-readiness]
- 104. Ten quick tips for biomarker discovery and validation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9371329/]
- 105. A novel disulfidptosis-related lncRNA signature for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12582836/]