High-Throughput Protein Crystallization Screens: A Modern Guide for Accelerated Drug Discovery and Structural Biology
This article provides a comprehensive guide to high-throughput protein crystallization, a critical technology for determining 3D protein structures in structural biology and rational drug design.
High-Throughput Protein Crystallization Screens: A Modern Guide for Accelerated Drug Discovery and Structural Biology
Abstract
This article provides a comprehensive guide to high-throughput protein crystallization, a critical technology for determining 3D protein structures in structural biology and rational drug design. It covers the foundational principles of crystallization and explores the automated platforms, reagents, and robotic systems that enable the rapid screening of thousands of conditions. The content details advanced optimization strategies for challenging targets like membrane proteins and synthesizes the latest trends, including the integration of AI for condition prediction and automated image analysis. Aimed at researchers, scientists, and drug development professionals, this guide serves as a strategic resource for implementing and optimizing efficient crystallization pipelines to accelerate biomedical research.
The Foundation of High-Throughput Crystallography: From Basic Principles to Market Drivers
Defining High-Throughput Protein Crystallization and Its Role in Structural Biology
High-Throughput Protein Crystallization (HTPC) is an automated, industrialized approach to crystallizing biological macromolecules. It leverages robotics, miniaturization, and parallel processing to rapidly set up and analyze thousands of crystallization trials simultaneously [1] [2]. This methodology was largely driven by the demands of structural genomics initiatives, such as the Protein Structure Initiative (PSI), which aimed to determine protein structures on a large scale [2] [3]. The primary goal of HTPC is to overcome the major bottleneck in macromolecular X-ray crystallography—obtaining diffraction-quality crystals—thereby accelerating structure-based drug design and fundamental biological research [1] [4].
Key Principles and Workflow
The underlying principle of HTPC is the statistical sampling of a multidimensional parameter space to identify initial "hit" conditions conducive to crystallization [2] [5]. Key parameters include the protein sample itself, precipitant type and concentration, pH, buffer species, temperature, and additives [6] [7]. Given the impracticality of exhaustive screening, HTPC employs strategically designed screens to efficiently probe this vast chemical landscape [5].
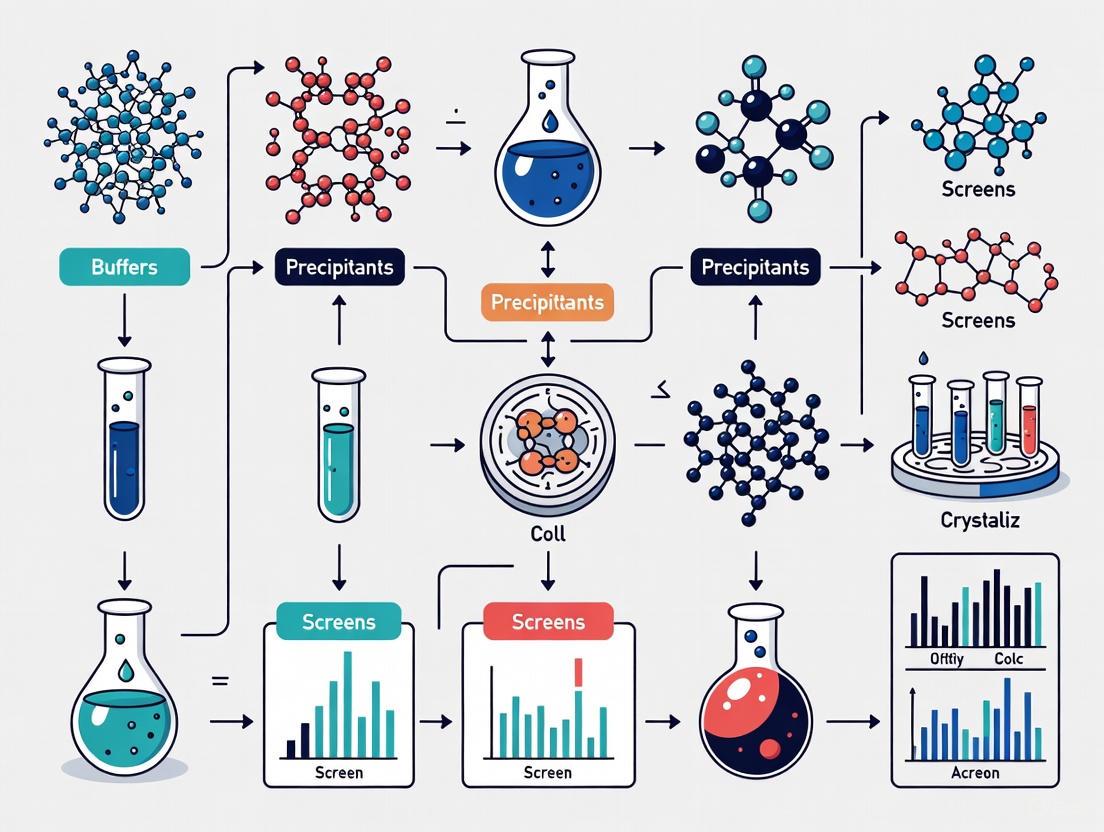
The following workflow diagram outlines the core stages of a high-throughput crystallization pipeline, from initial sample preparation to final structure determination.
- Sample Quality Control: The process begins with a purified protein sample. A critical prerequisite is ensuring the protein is homogeneous, monodisperse, and structurally intact, as sample purity is a major determinant of success [1] [2].
- Automated Screen Building: Liquid handling robots, such as the Formulator, are used to rapidly and precisely prepare crystallization screening solutions (cocktails) from a library of chemical stocks [8].
- Automated Trial Setup: Robotic systems like the NT8 Drop Setter dispense nanoliter-volume droplets of protein and screening solution, typically in sitting-drop, hanging-drop, or microbatch-under-oil formats [1] [8] [3]. This miniaturization allows thousands of trials to be performed with minimal protein consumption.
- Incubation & Automated Imaging: Plates are incubated under controlled temperature conditions and monitored by automated imaging systems (e.g., Rock Imagers) over days or weeks [8] [3].
- Image Analysis & AI Scoring: Advanced software and machine learning models (e.g., Sherlock) analyze the millions of images generated to classify outcomes and identify promising crystalline hits [8].
- Hit Identification & Optimization: Initial hits are used as starting points for optimization screens to produce larger, well-ordered crystals suitable for diffraction studies [6].
- X-ray Diffraction & Structure Determination: Optimized crystals are used for X-ray diffraction data collection, typically at synchrotron facilities, leading to three-dimensional structure determination [1] [3].
Key Methodologies and Experimental Protocols
Common Crystallization Techniques in HTPC
HTPC pipelines adapt common crystallization methods for automation and miniaturization. The table below compares the primary techniques used.
Table 1: Comparison of High-Throughput Crystallization Techniques
| Technique | Principle | Throughput & Automation | Protein Consumption | Key Applications in HTPC |
|---|---|---|---|---|
| Vapor Diffusion (Sitting/Hanging Drop) | Droplet containing protein and precipitant equilibrates via vapor phase against a larger reservoir solution [7]. | High; easily automated [6] [8]. | Small (nL-μL volumes) [1]. | Most widely used method for initial screening of soluble proteins [6] [7]. |
| Microbatch-under-Oil | A small batch droplet of protein and precipitant mixture is dispensed under an inert oil to prevent evaporation [6] [3]. | High; amenable to automation in 1536-well formats [3]. | Very small (nL volumes) [3]. | Used in large-scale screening centers (e.g., Hauptman-Woodward HTX Center); conditions are well-defined at setup [3]. |
| Lipidic Cubic Phase (LCP) | Protein is embedded in a lipidic mesophase, particularly suitable for membrane proteins [6]. | Possible with specialized robots [8]. | Small to Large [8]. | Crystallization of membrane proteins and GPCRs [6] [8]. |
| Free-Interface Diffusion | Protein and precipitant solutions diffuse into one another at a narrow interface within a capillary or microchannel [7]. | Difficult to automate [8]. | Very Small [8]. | Occasionally used for generating initial leads for difficult targets [7]. |
Protocol: High-Throughput Screening via Microbatch-under-Oil
This protocol is based on the established pipeline at the National High-Throughput Crystallization Center (HTX Center) [3].
I. Materials and Reagents
- Protein Sample: Purified, concentrated, and in a low-salt buffer. Goal: >150 μL at ≥ 5 mg/mL.
- Screening Solutions: Pre-configured 1536-condition screens for soluble or membrane proteins [3].
- Platform: 1536-well microassay plate.
- Sealing Agent: High-viscosity paraffin oil.
- Equipment: Automated liquid handling robots, automated imager.
II. Procedure
- Plate Preparation: Using a liquid-handling robot, dispense ~200 nL of each crystallization screening condition into the wells of a 1536-well microassay plate [3].
- Protein Dispensing: Dispense ~200 nL of the protein sample into each well, creating a microbatch droplet with the screening solution [3].
- Sealing: Immediately cover the entire plate with a layer of high-viscosity paraffin oil to prevent droplet evaporation [3].
- Incubation and Imaging:
- Seal the plate and incubate at a controlled temperature (e.g., 23°C or 14°C).
- Place the plate in an automated imaging system.
- Acquire images of all wells according to a predefined schedule (e.g., days 1, 3, 7, 14, 21, 28, 42) [3].
- Analysis: Use integrated software and AI-based autoscoring models to analyze the image dataset for the presence of crystals, precipitate, or other phase changes [8].
Protocol: Optimization from Initial Hits
Once a hit is identified, systematic optimization is crucial.
- Fine-Screening: Set up a fine-grid screen around the initial hit condition. Variations typically include:
- Precipitant Concentration: A gradient above and below the hit concentration.
- pH: A narrow pH range (e.g., ± 0.5 pH units) from the hit condition.
- Temperature: Replicate trials at multiple temperatures (e.g., 4°C, 20°C).
- Additives: Include small molecules, ions, or ligands that may enhance crystal order [6].
- Seeding: If crystals are numerous but small, use seeding techniques to transfer microscopic crystal fragments into new, slightly less saturated pre-equilibrated drops to promote larger crystal growth [6].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Research Reagent Solutions and Materials for HTPC
| Item | Function in HTPC | Examples / Notes |
|---|---|---|
| Sparse-Matrix Screens | Pre-mixed solutions that randomly sample crystallization chemical space based on historical success [6] [5]. | Hampton Research Crystal Screens, Molecular Dimensions JCSG+ screens. |
| Grid Screen Reagents | Individual stock solutions of precipitants (e.g., PEGs, salts), buffers, and additives for designing customized screens [5]. | Used with automated screen builders like the Formulator [8]. |
| Crystallization Plates | Microplates with wells designed for nanoliter-volume sitting or hanging drops, or microbatch experiments. | 96-well, 384-well, and 1536-well formats [1] [3]. |
| Paraffin Oil | An inert, high-viscosity oil used in microbatch-under-oil experiments to prevent evaporation of nanoliter droplets [3]. | Critical for the microbatch protocol at the HTX Center [3]. |
| Detergents | Essential for solubilizing and crystallizing membrane proteins by mimicking the lipid bilayer environment. | Included in specialized membrane protein screens (e.g., MembFac) [6] [3]. |
| Cryoprotectants | Chemicals (e.g., glycerol, ethylene glycol) added to crystals prior to flash-cooling in liquid nitrogen to prevent ice formation during X-ray data collection. | A standard consumable in downstream processing [9]. |
Quantitative Outcomes and Success Rates
The efficiency of HTPC is demonstrated by its scale and success metrics from operational centers.
Table 3: Quantitative Outcomes from High-Throughput Crystallization Screening
| Metric | Value | Context / Source |
|---|---|---|
| Screening Throughput | 40,000 experiments per day | Early automated system reported by [1]. |
| Drop Volume | 20 - 100 nL | Standard for nanodroplet HTPC robots [1]. |
| Standard Screen Size | 1,536 conditions | Standard screen size at the HTX Center [3]. |
| Crystal Hit Rate | 27% - 52% | Range of success rates for different PSI consortia at the HTX Center [3]. |
| Structure Determination Rate | 21% - 23.1% | Percentage of samples leading to a deposited PDB structure after optimization at the HTX Center [3]. |
| Total Samples Screened | >18,000 | Cumulative samples processed by the HTX Center over 20+ years [3]. |
HTPC has become a cornerstone of modern structure-based drug design [2] [4]. Its ability to rapidly determine the structures of protein-target complexes enables:
- Fragment-Based Drug Discovery (FBDD): Screening small, low-molecular-weight fragments that bind to a target, with HTPC providing structural information to guide their optimization into lead compounds [4].
- Lead Optimization: Providing atomic-level details of drug-target interactions, allowing medicinal chemists to rationally improve the potency and selectivity of drug candidates [2] [4].
In conclusion, High-Throughput Protein Crystallization has transformed crystallography from a specialized art into an industrialized, data-rich science. By integrating automation, miniaturization, and informatics, HTPC has significantly accelerated the pace of structural biology, providing indispensable insights for understanding biological mechanisms and developing new therapeutics.
The global protein crystallization market is demonstrating robust growth, propelled by its critical role in structural biology and drug discovery. The market is poised to expand from $1.62 billion in 2024 to $2.8 billion by 2029, reflecting a strong Compound Annual Growth Rate (CAGR) of 11.5% [9] [10]. This trajectory is supported by rising R&D investments and an increasing focus on protein-based therapeutics.
Table 1: Global Protein Crystallization Market Financial Projection
| Metric | 2024 Value | 2029 Projection | CAGR |
|---|---|---|---|
| Market Size | $1.62 Billion [9] [10] | $2.8 Billion [9] [10] | 11.5% [9] |
Market Segmentation Analysis
Growth is pervasive across all market segments, with specific areas showing accelerated adoption driven by technological advancements.
Table 2: Protein Crystallization Market Size and Growth by Segment
| Segmentation | Dominant Segment (2024) | Fastest-Growing Segment | Key Growth Driver |
|---|---|---|---|
| Product | Consumables [11] [12] [13] | Software & Services (12.19% CAGR) [14] | High-throughput screening demands and AI-driven data analysis [14]. |
| Technology | X-ray Crystallography [14] [13] | Microfluidic Chip-Based Screening (11.73% CAGR) [14] | Miniaturization, reduced sample volume, and faster results [14]. |
| End User | Pharmaceutical & Biotechnology Companies [14] | Contract Research Organizations (CROs) (10.24% CAGR) [14] | Outsourcing of specialized crystallization workflows [14]. |
| Region | North America (36.13% share) [14] | Asia-Pacific (10.05% CAGR) [14] | Strong R&D infrastructure and major biopharma presence [11]; rising investments and pharmaceutical expansion [13]. |
Key Growth Factors: An Analytical Framework for High-Throughput Research
The market's expansion is underpinned by several interconnected factors that directly influence high-throughput screening (HTS) methodologies.
Primary Market Drivers
- Escalating Demand for Biopharmaceuticals: The shift towards targeted protein therapeutics, including monoclonal antibodies and engineered enzymes, requires atomic-level structural information for rational drug design. Protein crystallization is indispensable for providing this structural insight, refining targeted therapies for better efficacy and stability [10] [14].
- Increased R&D Investment: Significant funding from both public and private sectors is accelerating structural biology research. For instance, the Australian government invested $4.34 billion in R&D in 2022-23, a substantial increase from previous years [9] [10]. Such investments legitimize capital expenditures on advanced crystallization platforms [14].
- Technological Convergence and Automation: The integration of automation, robotics, and Artificial Intelligence (AI) is transforming traditional workflows. AI algorithms predict optimal crystallization conditions, while robotic liquid handling systems enable high-throughput screening of thousands of conditions, drastically improving efficiency and success rates [9] [14] [15].
Pivotal Market Trends
- Miniaturization via Microfluidics: Microfluidic devices and lab-on-a-chip technologies are revolutionizing HTS by allowing experiments with nanoliter-volume samples. This reduces reagent consumption and protein sample requirements, which is particularly valuable for studying difficult-to-express proteins [14] [16].
- Adoption of Hybrid Structural Biology Techniques: Cryo-electron Microscopy (Cryo-EM) is increasingly used alongside traditional X-ray crystallography. Cryo-EM provides high-resolution structural information for proteins that are challenging to crystallize, creating complementary workflows in drug discovery pipelines [11] [14].
- Innovative Crystallization Methods: Novel techniques like cell-free protein crystallization, developed by the Tokyo Institute of Technology, enable the study of unstable proteins that are intractable with conventional methods, opening new frontiers in structural biology [9] [10].
Application Notes: High-Throughput Screening Protocol for Membrane Proteins
The following protocol provides a detailed methodology for high-throughput crystallization screening of challenging targets like membrane proteins, incorporating contemporary technologies and reagents.
Experimental Workflow: High-Throughput Crystallization Screening
The diagram below outlines the key stages of a modern, high-throughput crystallization screening workflow.
Protocol Steps
Protein Sample Preparation
- Aim: Obtain a pure, monodisperse, and concentrated protein sample.
- Procedure:
- Purify the target membrane protein using affinity and size-exclusion chromatography in a suitable detergent.
- Use analytical size-exclusion chromatography to confirm monodispersity.
- Concentrate the protein to ≥ 40 mg/mL in a compatible buffer using centrifugal concentrators.
- Centrifuge at 14,000 × g for 20 minutes at 4°C to remove any aggregates immediately before setting up crystallization trials.
Primary High-Throughput Screening
- Aim: Identify initial crystallization "hits" from a broad spectrum of conditions.
- Procedure:
- Use an automated liquid handling robot (e.g., Mosquito LCP or Dragonfly).
- Dispense 150-200 nL of protein solution per well in a 96-well sitting-drop crystallization plate.
- For membrane proteins, consider using the Lipidic Cubic Phase (LCP) method with the same robot.
- Dispense 100 nL of a commercial sparse matrix screen (e.g., MemGold1/2, MemMeso) as the precipitant solution.
- Seal the plate with a transparent tape and incubate at a controlled temperature (e.g., 20°C).
Automated Imaging and Crystal Detection
- Aim: Monitor plates and identify crystal formation without manual intervention.
- Procedure:
- Load plates into an automated imaging system (e.g., Formulatrix Rock Imager series).
- Program the imager to scan each well every 6-8 hours for the first 7 days, then daily for up to 90 days.
- Use integrated AI-based image analysis software to automatically score wells and flag potential crystalline hits based on predefined morphological features.
Hit Optimization
- Aim: Refine initial hits to produce large, diffraction-quality crystals.
- Procedure:
- Prepare a fine-screen around the chemical space of the initial hit condition (e.g., varying pH ± 0.5, precipitant concentration ± 20%).
- Employ microseed screening (MSS) to improve crystal size and uniformity.
- Prepare a seed stock by crushing initial microcrystals.
- Add a diluted seed stock to each new crystallization drop to nucleate growth.
Crystal Harvesting and Data Collection
- Aim: Harvest and freeze crystals for high-resolution X-ray diffraction.
- Procedure:
- Harvest optimized crystals using micromount loops (e.g., MiTeGen MicroLoops).
- Cryo-cool crystals by plunging into liquid nitrogen after a brief soak in a cryoprotectant solution.
- Ship crystals to a synchrotron facility under liquid nitrogen for high-brilliance X-ray exposure.
- Collect a complete dataset, typically 180-360 degrees of rotation.
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful high-throughput crystallization screens rely on a suite of specialized reagents and instruments.
Table 3: Essential Research Reagent Solutions for High-Throughput Crystallization
| Item Category | Specific Product Examples | Function in Experiment |
|---|---|---|
| Crystallization Plates | Corning Next Generation CrystalEX Microplates [13], Greiner Bio-One 96-well sitting-drop plates | Provide a miniaturized platform for setting up thousands of vapor-diffusion trials with nanoliter volumes. |
| Sparse Matrix Screens | Hampton Research Index, Jena Bioscience JBScreen, Molecular Dimensions MemGold/MemMeso | Pre-formulated suites of conditions that sample a diverse chemical space (precipitants, salts, buffers, additives) to identify initial crystallization hits. |
| Liquid Handling Instruments | SPT Labtech mosquito, TTP Labtech dragonfly | Automated robots capable of precisely dispensing nanoliter volumes of protein and precipitant, enabling high-throughput, reproducible plate setup. |
| Automated Imaging Systems | Formulatrix Rock Imager 8S [12] | High-throughput microscopes that automatically image crystallization drops at scheduled intervals, allowing for kinetic monitoring of crystal growth. |
| AI-Powered Analysis Software | ROCK MAKER (Formulatrix) [13], AI-based crystal detection algorithms [9] | Software that uses machine learning to analyze images from imaging systems, automatically identifying and scoring potential crystals based on morphology. |
The protein crystallization market's trajectory toward $2.8 billion by 2029 is intrinsically linked to technological advancements that address the core challenges of high-throughput screening. The integration of automation, miniaturization, and artificial intelligence is creating a new paradigm where obtaining structural information is faster, more efficient, and more accessible. For researchers and drug development professionals, leveraging the protocols, tools, and insights outlined in these application notes is crucial for staying at the forefront of structural biology and accelerating the development of next-generation therapeutics.
Building a Modern HT Crystallization Pipeline: Automation, Reagents, and Techniques
Within structural biology and rational drug design, determining the three-dimensional structure of proteins is paramount for understanding function and guiding the development of therapeutic molecules. X-ray crystallography, the predominant method for structure determination, requires high-quality, well-ordered single crystals [17]. The production of these crystals often represents the most significant bottleneck in the entire pipeline [6] [17]. Protein crystallization is a complex multiparametric process, and finding initial "hit" conditions has been compared to searching for a "needle in a haystack" [6].
The advent of high-throughput methodologies has dramatically accelerated the process of crystallization screening, cutting the setup and analysis time from weeks to minutes and enabling the testing of thousands of conditions [6]. Among the plethora of techniques developed, vapor diffusion, microbatch, and lipid cubic phase (LCP) crystallization have emerged as critical methods. Vapor diffusion remains the most widely used technique, while LCP has revolutionized the crystallization of membrane proteins, a class of targets constituting over 60% of modern drugs [18]. This application note provides a detailed comparison of these three pivotal methods, offering structured data and actionable protocols to support researchers in selecting and implementing the optimal crystallization strategy for their specific projects.
Theoretical Principles and Comparative Analysis
Fundamental Crystallization Thermodynamics
All protein crystallization methods share a common goal: to gently drive a purified protein solution into a state of supersaturation, which is the thermodynamic driving force for nucleation and subsequent crystal growth [19] [17] [20]. A typical protein phase diagram can be divided into regions of undersaturation, metastability, and labile (supersaturated) zones [19]. The optimal path navigates through a narrow window of supersaturation that is high enough to promote nucleation but low enough to avoid amorphous precipitation [19]. The different methods achieve this supersaturation through distinct physical mechanisms, which directly influence their success rates, crystal quality, and applicability.
Method Comparison Table
The following table summarizes the key characteristics, advantages, and challenges of vapor diffusion, microbatch, and LCP crystallization.
| Feature | Vapor Diffusion | Microbatch | Lipid Cubic Phase (LCP) |
|---|---|---|---|
| Primary Principle | Equilibration of a droplet against a reservoir via vapor phase, concentrating the sample [17] [21]. | Direct mixing of protein and precipitant under oil, preventing evaporation [17]. | Crystallization within a lipidic mesophase that mimics the native membrane environment [18]. |
| Typely Protein Consumption | Small to Large (e.g., 2-4 μL drops) [8] | Small (e.g., 0.5-1 μL drops) [22] | Small to Large [8] |
| Throughput & Automation | High; easily automated with liquid handlers [8] [6]. | High; suitable for robotics (e.g., IMPAX machine) [22]. | Possible; specialized robots required [8]. |
| Speed of Nucleation | Relatively fast; conditions change dynamically [22]. | Variable; can be immediate or slow, depending on formulation [22]. | Typically slower; occurs within the mesophase matrix [18]. |
| Control Over Supersaturation | High, through reservoir composition and drop ratio [21]. | Fixed at setup; can be modified with oil mixtures for evaporation [22] [23]. | High, but complex; depends on lipid composition and precipitant diffusion [18]. |
| Crystal Quality & Size | Can produce large crystals; quality can be excellent [17]. | Crystals may be smaller than in VD under the same condition [22]. | Often produces high-quality, well-ordered crystals for membrane proteins [18]. |
| Seeding & Handling | Possible and relatively straightforward [8]. | Not possible in traditional setup [8]. | Possible, though harvesting can be difficult [8]. |
| Ideal Application | Broad-purpose, initial screening, and optimization [8] [21]. | Rapid screening, low protein availability, and specific optimization [22] [23]. | Membrane proteins, integral membrane proteins [8] [18]. |
| Key Challenges | Sensitive to environmental fluctuations, longer equilibration times [20]. | Wide crystal size distribution possible, less dynamic control [20]. | Technically challenging, harvesting can be difficult, limited to specific protein types [8] [18]. |
Quantitative Performance Insights
A comparative study screening six proteins with sparse matrix conditions found that a combination of three microbatch variants identified 43 out of 58 total conditions (74%), while a single vapor diffusion screen identified 41 conditions (71%) [22]. Critically, each method discovered unique hits: approximately 29% of conditions would have been missed if microbatch had not been employed alongside vapor diffusion [22]. Furthermore, vapor diffusion typically produces crystals more quickly in the initial weeks, whereas microbatch (particularly an evaporation variant) continues to produce new conditions over longer durations, eventually matching vapor diffusion's total yield [22].
Experimental Protocols
Vapor Diffusion Protocol (Sitting Drop)
Principle: A droplet containing a mixture of protein and precipitant is sealed in a chamber with a pure precipitant reservoir. Water vapor diffuses from the drop to the reservoir, slowly concentrating the protein and precipitant until equilibrium is achieved, ideally crossing the supersaturation threshold for nucleation [17] [21].
Materials:
- Purified, concentrated protein (>99% pure, typically 5-50 mg/mL) [17] [21]
- 24-well sitting drop tray (e.g., from Hampton Research)
- Reservoir solutions (e.g., commercial sparse matrix screens)
- Optically clear sealing tape
- Low-retention pipette tips (0.1-2 μL)
- Siliconized glass cover slides (for hanging drop variant)
Procedure:
- Preparation: Filter all stock solutions using a 0.22 μm filter. Centrifuge the protein sample (e.g., 15 min at 18,000 x g, 4°C) to remove any aggregates [17].
- Reservoir Setup: Pipette 500 μL of precipitant reservoir solution into each well of the crystallization tray [17].
- Drop Dispensing: On the raised shelf of the sitting drop well, combine 1 μL of protein solution with 1 μL of reservoir solution. Mix carefully by pipetting, avoiding bubble formation [17].
- Sealing: Use optically clear sealing tape to securely cover the entire tray, ensuring a complete vapor-tight seal for each well.
- Incubation: Place the tray gently on a stable, vibration-damped surface in a temperature-controlled incubator (commonly 4°C or 20°C). Avoid disturbances [17].
- Monitoring: Check the drops for crystal formation the following day, and then regularly every few days using a microscope. Document all outcomes [17].
Microbatch Crystallization Protocol
Principle: Protein and precipitant solutions are directly mixed in a single step and dispensed under an oil layer, which prevents evaporation and fixes the crystallization condition from the outset [17]. A modified version uses a specific oil mixture to allow slow water evaporation, mimicking vapor diffusion [22] [23].
Materials:
- Purified, concentrated protein
- 96-well microbatch tray (e.g., Nunc HLA plates)
- Paraffin oil (e.g., from Sigma-Aldrich)
- Silicone oil (for evaporation method)
- Liquid handling robot (e.g., Douglas Instruments IMPAX) or manual pipettes with fine tips
Procedure:
- Tray Preparation: Clean the microbatch tray with compressed air to remove dust. Fill the wells with paraffin oil to a depth of approximately 3 mm [17].
- Droplet Formation (Under Oil):
- For standard microbatch: Pipette 1 μL of protein solution directly to the bottom of an oil-filled well. Add 1 μL of precipitant solution to the same well, ensuring the droplets sink and fuse at the bottom [17].
- For evaporation-controlled microbatch: Use a 50:50 mixture of paraffin and silicone oil. The silicone oil permeability allows slow water evaporation, gradually increasing concentration over time [22].
- Incubation and Monitoring: Seal the plate (if necessary) and incubate as described for vapor diffusion. Monitor regularly for crystal growth [22].
Lipid Cubic Phase (LCP) Crystallization Protocol
Principle: The membrane protein is reconstituted into a lipidic cubic mesophase, a semi-solid matrix of continuous lipid bilayers. Precipitant solution is then introduced, often in the form of an LCP "bolus" covered by a precipitant solution, and crystals grow within the lipid bilayer environment [18].
Materials:
- Purified membrane protein in a suitable detergent
- Lipids for mesophase formation (e.g., Monoolein)
- Precipitant solutions
- Syringe mixing setup (two gastight syringes connected by a coupler)
- LCP crystallization plates (e.g., 96-well glass sandwich plates)
Procedure:
- Mesophase Preparation: Mix Monoolein and the membrane protein solution (typically at a 60:40, w:w ratio) using two syringes connected by a coupler. Push the plungers back and forth repeatedly until the mixture forms a clear, viscous mesophase [18].
- Bolus Dispensing: Using a syringe, dispense 50-100 nL boluses of the protein-laden mesophase onto the well of an LCP crystallization plate.
- Precipitant Overlay: Carefully overlay each mesophase bolus with 1 μL of precipitant solution.
- Sealing and Incubation: Seal the plate with a glass cover slide and incubate. Crystals typically grow within the mesophase and can be detected as birefringent objects under a microscope [18].
- Harvesting: Harvesting crystals from LCP is complex and often involves using special MicroMounts (e.g., MiTeGen MicroLoops) to scoop out the entire bolus.
The Scientist's Toolkit: Essential Research Reagents and Materials
| Item | Function/Description | Example Suppliers/Notes |
|---|---|---|
| Precipitants | Reduce protein solubility to induce supersaturation. Polyethylene glycol (PEG) and ammonium sulfate are the most common, accounting for ~60% of successful conditions [17] [20]. | Hampton Research, Molecular Dimensions |
| Sparse Matrix Screens | Pre-formulated screening solutions based on historical crystallization data, allowing efficient exploration of chemical space [6] [17]. | Crystal Screen (Hampton Research), JCSG Core (Molecular Dimensions) |
| Crystallization Plates | Specialized plates with wells for reservoir solutions and platforms for drop deposition. | 24-well Vapor Diffusion Plates, 96-well Sitting Drop Plates, LCP Glass Sandwich Plates |
| Liquid Handling Robots | Automate the dispensing of nanoliter-volume drops, ensuring precision, reproducibility, and high throughput [8] [6]. | NT8 Drop Setter (Formulatrix), IMPAX (Douglas Instruments) |
| Lipids for LCP | Form the bicontinuous cubic phase matrix that hosts membrane proteins. Monoolein is a standard lipid used [18]. | Nu-Chek Prep |
| Automated Imaging Systems | Capture high-quality images of crystallization drops over time for remote monitoring and analysis [8]. | Rock Imager series (Formulatrix) |
| Laboratory Information Management System (LIMS) | Software to manage the entire crystallization workflow, from plate setup and tracking to image analysis and data management [8]. | Rock Maker (Formulatrix) |
Method Selection Workflow
The following diagram outlines a logical decision pathway for selecting the most appropriate crystallization method based on key project parameters.
The strategic selection of a crystallization method is a critical determinant of success in high-throughput structural biology pipelines. As demonstrated, vapor diffusion, microbatch, and LCP are not mutually exclusive but are complementary tools, each with distinct strengths. Vapor diffusion offers dynamic control and is the workhorse for soluble proteins. Microbatch conserves precious sample and can uncover unique conditions. LCP is indispensable for tackling the challenging landscape of membrane protein structural biology. The integration of these methods, guided by rational decision-making and supported by automation and sophisticated data management, provides a powerful framework for advancing research in structural biology and drug development. Future directions will likely involve greater integration of machine learning for predictive modeling and condition optimization, further enhancing the efficiency and success of protein crystallization endeavors [8].
In the field of structural biology and rational drug discovery, high-throughput protein crystallography has become an indispensable technology for determining the three-dimensional structures of biological macromolecules [2]. The process of obtaining diffraction-quality crystals remains a significant bottleneck, compounded by the fact that crystallization conditions for novel samples cannot be predicted [24]. Automated workflows address this challenge by enabling the rapid, reproducible, and systematic screening of thousands of crystallization conditions while conserving precious protein samples [25] [26]. This application note details integrated protocols for automating the key stages of protein crystallization: screen building, drop setting, and automated imaging, providing a framework for laboratories aiming to implement or enhance high-throughput structural biology pipelines.
The Integrated Automated Workflow
The transition from manual to automated crystallization involves the seamless integration of specialized instrumentation, software, and standardized protocols. The entire process, from designing the experiment to analyzing the results, can be managed within a unified system. The workflow below illustrates the core pathway and the technologies involved at each stage.
Equipment and Reagent Solutions
A robust automated crystallization platform relies on integrated hardware and software components. The table below summarizes key solutions for establishing this workflow.
Table 1: Essential Research Reagent Solutions and Instrumentation for Automated Protein Crystallization
| Category | Product/System Name | Key Function | Technical Specifications |
|---|---|---|---|
| Laboratory Information Management System (LIMS) | Rock Maker [25] [8] | Manages the entire experimentation process, from experimental design and dispense to image viewing and analysis. | Integrates with screen builders, drop setters, and imagers; provides tools for AI-based autoscoring. |
| Screen Builder | Formulator Screen Builder [25] [8] | Prepares crystallization screening solutions by dispensing up to 34 different ingredients. | Uses a 96-nozzle dispensing chip; volumes from 200 nL; no consumables required. |
| Drop Setter / Crystallization Robot | NT8 Drop Setter [25] [8] | Sets up crystallization experiments (hanging/sitting drop, LCP, etc.) by combining protein and screen solutions. | 8-tip head; dispenses drops from 10 nL to 1.5 μL; features active humidification and reusable tips. |
| Automated Imager | Rock Imager Series [25] [8] | Automated imaging system for crystallization plates with various capacities and advanced imaging modalities. | Models available with capacities from 1 to 1000 plates; imaging options include Visible, UV, MFI, and SONICC. |
| Alternative Liquid Handler | Opentrons-2 [27] | An economical and versatile liquid handling robot for automating crystallization plate setup, controlled via Python. | Accessible platform for mixing and setting up 24-well sitting drop vapor diffusion trials. |
Detailed Protocols
Protocol 1: Fully Automated Preparation of Crystallization Screening Plates
This protocol describes the creation of a large stock of pre-dispensed screening plates (e.g., "LMB plates") for use in initial crystallization trials [24]. This system integrates a liquid handler, an automated carousel, an inkjet printer, and an adhesive plate sealer.
Materials:
- Formulatrix Formulator Screen Builder or equivalent [25]
- Commercially available screening kit solutions
- SBS-standard 96-well crystallization plates
- Adhesive sealing films
- Laboratory Information Management System (LIMS)
Procedure:
- System Initialization: Ensure the liquid handler and plate sealer are initialized and software is running. Activate the tube-cooling carrier chiller 30 minutes prior to start [24].
- Plate Sealer Test: Place a test plate on the sealer carrier and run the sealing process three times to verify the film is applied correctly [24].
- Liquid Handler Priming: Fill the main container of the liquid handler with deionized water and connect the fluidic coupling. Enter the screen name on the inkjet printer and initiate carousel rotation [24].
- Plate and Reagent Loading:
- Liquid Handler Flushing: Run a maintenance flush program to prime the system and wash the dispensing tips [24].
- Plate Dispensing, Labeling, and Sealing:
- Execute the "MRC kit dispensing" program. The system will automatically [24]:
- Transfer 80 µL of each screening condition from the source tubes to the reservoir wells of the crystallization plates.
- Inkjet-print a unique identifier on each plate.
- Apply an adhesive seal to each finished plate.
- Execute the "MRC kit dispensing" program. The system will automatically [24]:
- System Shutdown and Plate Storage: After all plates are processed, switch the liquid handler to an ethanol rinsing solution and run a flush program. Turn off the chiller. Store the successfully prepared plates at a constant temperature (e.g., 10°C) [24].
Protocol 2: High-Throughput Crystallization Setup Using a Drop Setter
This protocol utilizes a dedicated crystallization robot to set up 1,920 initial screening conditions for a single protein sample across twenty 96-well plates via sitting drop vapor diffusion [24].
Materials:
- Purified protein sample (> 2 mg/mL concentration) [24] [2]
- Pre-dispensed screening plates (from Protocol 1)
- NT8 Drop Setter or equivalent nanoliter dispenser [25] [8]
- SBS-standard sitting drop crystallization plates (if not using pre-dispensed ones)
Procedure:
- System Preparation: Power on and initialize the nanoliter dispenser and liquid handler. Activate the microtube-cooling carrier 15 minutes beforehand. Test the plate sealer and the nanoliter dispenser's droplet formation using a test solution [24].
- Plate Loading: Insert the first pre-dispensed screening plate into the custom plate holder on the deck and remove its adhesive seal. Place the plate on the deck's sliding carrier [24].
- Sample Loading: Centrifuge the purified protein sample briefly and place it in the cooling carrier on the deck.
- Automated Drop Setting:
- The liquid handler and nanoliter dispenser work in concert to [24] [8]:
- Combine 100 nL of the protein sample with 100 nL of the crystallization condition from the reservoir.
- Dispense this 200 nL mixed droplet onto the sitting drop post or shelf.
- This process is repeated for all 96 wells of the plate.
- The liquid handler and nanoliter dispenser work in concert to [24] [8]:
- Plate Sealing and Storage: Once all droplets are set, the robotic system automatically applies a transparent adhesive seal to the plate. The plate is then transferred to a temperature-controlled incubator or storage hotel [24].
- Process Repetition: The system proceeds to set up the remaining 19 plates automatically, creating a total of 1,920 individual crystallization trials for the sample [24].
Protocol 3: Automated Imaging and Hit Detection
This protocol covers the automated monitoring of crystallization trials and identification of crystal hits using advanced imaging technologies.
Materials:
Procedure:
- Plate Scheduling: Within the LIMS (e.g., Rock Maker), define an imaging schedule for the batch of crystallization plates. Specify the frequency of imaging (e.g., daily for the first week, weekly thereafter) and the imaging modalities to be used [26].
- Automated Plate Retrieval and Imaging: The automated imager, potentially integrated with a plate hotel, will [25] [28]:
- Retrieve plates from storage according to the schedule.
- Position each well under the optics automatically.
- Capture images using pre-defined modalities (see Table 2).
- Return the plate to its storage location.
- Image Analysis:
- Manual Review: Acquired images are stored in a central database. Users can remotely view and score the experiments for crystal growth, precipitation, or phase separation [26].
- AI-Assisted Scoring: Utilize integrated AI autoscoring models (e.g., MARCO or Sherlock) to pre-analyze the images. These models can highlight potential hits, significantly reducing the time required for manual inspection [8].
Table 2: Imaging Modalities for Crystal Hit Identification
| Imaging Modality | Principle | Application | Key Advantage |
|---|---|---|---|
| Visible Light [8] | Bright-field, dark-field, or phase-contrast microscopy using visible spectrum light. | General monitoring of crystal growth and drop morphology. | Suitable for analyzing large, well-formed crystals. |
| Ultraviolet (UV) [8] [28] | Excitation of intrinsic fluorescence from aromatic amino acids (e.g., Tryptophan). | Distinguishing protein crystals from salt crystals. | Label-free method for confirming the proteinaceous nature of a crystal. |
| Second Order Nonlinear Imaging of Chiral Crystals (SONICC) [25] [8] | Combines Second Harmonic Generation (SHG) and UV-TPEF. | Detecting microcrystals (<1 μm) and crystals obscured in precipitate or LCP. | High sensitivity and ability to definitively identify protein crystals. |
| Multi-Fluorescence Imaging (MFI) [8] [28] | Detection of fluorescence from covalently attached fluorophores (Trace Fluorescent Labeling). | Identifying protein crystals in complex mixtures and distinguishing them from salt. | High sensitivity, especially when intrinsic protein fluorescence is weak. |
The implementation of a fully automated workflow from screen building to automated imaging represents a paradigm shift in macromolecular crystallography. It directly addresses the critical bottlenecks of precision, reproducibility, and throughput that challenge crystallography laboratories [29] [26]. By standardizing liquid dispensing with nanoliter accuracy, these systems drastically reduce consumption of valuable protein samples, allowing for more extensive screening of chemical space [2] [26]. Furthermore, integration with a LIMS ensures rigorous tracking of experimental parameters, enabling data-driven optimization and enhancing the overall reliability of the structural pipeline.
While high-throughput approaches are powerful, they do not replace the need for expert analysis. The success of these automated systems ultimately relies on skilled researchers to interpret results, particularly complex crystallization outcomes, and to design intelligent optimization strategies based on the initial screening data [2]. The protocols outlined herein provide a foundation for laboratories to achieve a high level of automation, accelerating the path from protein sample to diffraction-quality crystal and, ultimately, to a three-dimensional structure that can inform biological understanding and drug discovery efforts.
Protein crystallization is a critical process in structural biology, forming a regular array of individual protein molecules stabilized by crystal contacts. Understanding protein structure is of great importance for predicting function, studying protein-protein or ligand-protein interactions for drug discovery, and uncovering stages in enzyme catalysis [8]. The process involves gradually reducing the solubility of the protein using precipitants in a controlled environment, but predicting optimal conditions remains challenging due to the complex interplay of interactions between protein molecules [8]. Automation has become the definitive response to overcoming the crystallization bottleneck in biological crystallography, transforming a traditionally slow, resource-intensive, and error-prone process into a streamlined, reproducible, and high-throughput workflow [8] [30]. This application note details the essential tools and methodologies for implementing automated protein crystallization within the context of high-throughput research, providing structured protocols for researchers, scientists, and drug development professionals.
The transition to automated platforms addresses several critical limitations of manual methods. Conventional protein crystallization workflows involve working with sub-microliter volumes, where even slight inaccuracies in dispensing or mixing reagents lead to significant errors or suboptimal results [8]. Automation eliminates human error, ensures reproducibility, and dramatically increases throughput by integrating and standardizing all crystallization steps [8]. High-throughput facilities are now capable of setting up thousands of crystallization trials per day, testing multiple constructs of each target on a production-line basis, which has significantly improved success rates and made crystallization much more convenient [30].
Essential Equipment for High-Throughput Crystallization
A complete automated protein crystallization workflow integrates several specialized instruments: liquid handlers for preparing crystallization screens and setting up drops, crystallization robots capable of handling various techniques and sample types, and automated storage imagers for temperature-controlled incubation and visual inspection. The synergy between these components creates a seamless pipeline from experimental design to crystal detection.
Crystallization Robots and Liquid Handlers
Crystallization robots and liquid handlers form the core of automated setup, enabling precise, nanoliter-volume dispensing that conserves precious protein samples. These systems provide the accuracy and reproducibility essential for high-throughput screening.
Table 1: Comparison of Automated Crystallization Robots and Liquid Handlers
| Device Name | Type | Volume Range | Key Features | Supported Methods | Throughput |
|---|---|---|---|---|---|
| NT8 Drop Setter [8] | Liquid Handler (Tip-based) | 10 nL - 1.5 μL | Proportionally-controlled active humidification; reusable tips; 8-tip head | Sitting drop, hanging drop, LCP, microbatch, additives, seeding | Fast nanoliter-volume dispensing |
| Mosquito Crystal [31] | Liquid Handler (Tip-based) | 25 nL - 1.2 μL | "Walk up and use" technology; active humidity chamber; true positive displacement pipetting | Sitting drop, hanging drop, microbatch, seeding, additive screening | 2 minutes per 96-well plate |
| Formulator [8] | Screen Builder (Tipless) | 200 nL - no upper limit | Microfluidic dispenser; 96-nozzle chip; dispenses 34 different ingredients | Creation of crystallization screens | 2.7 minutes for a 96-well grid screen |
| Dragonfly Crystal [32] | Liquid Handler (Non-contact) | 0.5 μL - 4 mL | 10 independent dispensing heads; non-contact; contamination-free | Protein crystallization setup, assay plate preparation | 4-8 minutes per 96-well plate |
Automated Storage and Imaging Systems
After automated setup, crystallization plates require temperature-controlled incubation and regular monitoring. Automated storage imagers combine precise environmental control with advanced imaging capabilities to track crystal growth over time, identifying critical hits among thousands of experiments.
Table 2: Comparison of Automated Storage and Imaging Systems
| System Name/Type | Plate Capacity | Temperature Control | Imaging Modalities | Key Features |
|---|---|---|---|---|
| Rock Imager 1000 [8] [33] | 1000 plates | Yes (4°C - 30°C) | Visible, UV, MFI, SONICC | Highest capacity; floor-standing; full imaging options |
| Rock Imager 360 [8] [33] | 364 plates | Yes (4°C - 30°C) | Visible, UV, MFI | Benchtop; browser-based software; compact storage |
| Rock Imager 2 [8] | 2 plates | No | Visible, UV, MFI | Benchtop; Windows-based software |
| Rock Imager 1 [8] | 1 plate | No | Visible, UV | Basic benchtop model |
| SpectroQ UV [34] | Up to 690 SBS plates | Yes (4°C - 20°C) | Ultraviolet, Visible | Patented LED light source; high-resolution imaging |
Research Reagent Solutions
Successful high-throughput crystallization screening requires careful selection of reagents and materials. The following table outlines essential solutions and their specific functions in the experimental workflow.
Table 3: Essential Research Reagent Solutions for Automated Crystallization
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Precipitant Solutions [8] | Reduce protein solubility to promote crystallization | Hundreds of inorganic and organic compounds are used; systematically combined in screens |
| Crystallization Screens [8] [30] | Pre-formulated condition matrices for initial screening | Commercial screens available; can be custom-built using instruments like the Formulator |
| Lipidic Cubic Phase (LCP) Materials [8] | Matrix for crystallizing membrane proteins | Specialized protocol; requires compatible equipment (e.g., NT8 or Xantus robot) |
| Amine-Reactive Dyes [8] | Fluorescent labeling for Multi-Fluorescence Imaging (MFI) | Used to distinguish protein-protein complexes; choice of dye concentration is critical for stability |
| Buffers and Salts [8] [30] | Control pH and ionic strength of crystallization environment | Critical parameters systematically varied in screening; stock solutions prepared as concentrates |
| Crystallization Plates [33] [31] | Physical platform for experiments | Various formats: SBS, Linbro, Nextal, LCP; must match instrument specifications |
Experimental Protocols and Workflows
Comprehensive Workflow for High-Throughput Crystallization
The automated protein crystallization process follows a defined multi-step workflow that integrates instrumentation, software, and reagents. The diagram below illustrates this comprehensive pipeline from sample preparation to final analysis.
Protocol 1: Automated Initial Screening Setup
Purpose: To efficiently screen a purified protein sample against a broad matrix of crystallization conditions using automated liquid handling.
Materials:
- Purified protein sample (>95% purity, concentrated)
- Commercial crystallization screen solutions
- Suitable crystallization plates (SBS format)
- NT8 Drop Setter or Mosquito Crystal liquid handler
- Rock Maker software
Procedure:
- Experimental Design: In Rock Maker software, design the screening experiment by selecting the desired commercial screen and plate type. Define the drop ratio (typically 1:1, 2:1, or 3:1 protein:precipitant) and volume (50-200 nL total) [8] [35].
- Plate Preparation: Using the Formulator screen builder or equivalent, dispense crystallization screen solutions into the plate reservoirs according to the selected screen [8].
- Protein Dispensing: Program the liquid handler (NT8 or Mosquito Crystal) to dispense nanoliter volumes of protein and precipitant solutions according to the designed experiment. For sitting drop vapor diffusion, typical drop sizes range from 50 nL to 400 nL [8] [31].
- Sealing and Transfer: Automatically seal the plate and transfer it to the automated storage imager. The entire process from experimental design to plate setup is coordinated through Rock Maker software [35].
Critical Notes: Maintain active humidification during drop setup to prevent evaporation, especially with nanoliter volumes [8] [31]. Include control drops when possible. Protein concentration should be optimized prior to screening (typically 5-20 mg/mL).
Protocol 2: Automated Imaging and Crystal Detection
Purpose: To automatically monitor, image, and identify crystal formation in high-throughput crystallization trials over time.
Materials:
- Rock Imager or SpectroQ UV system with temperature control
- Crystallization plates from Protocol 1
- Rock Maker software with AI-based autoscoring
Procedure:
- Plate Registration: Register the crystallizations plates in the Rock Imager system using barcode identification, which links physical plates to the experimental design in Rock Maker [36].
- Imaging Schedule: Define an automated imaging schedule in Rock Maker software. Typical schedules image plates immediately after setup (day 0), then daily for the first week, and progressively less frequently over weeks to months [8] [33].
- Multi-Modal Imaging: Configure the imager to capture images using multiple modalities as required:
- Automated Analysis: Enable AI-based autoscoring models (MARCO or Sherlock) integrated with Rock Maker to analyze image datasets and identify potential hits [8].
Critical Notes: UV imaging requires protein crystals containing tryptophan residues. For membrane proteins in LCP, FRAP (Fluorescence Recovery After Photobleaching) can pre-screen conditions without waiting for crystal growth [8]. Regular calibration of imaging systems is essential for consistent results.
Advanced Applications and Techniques
Imaging Modalities for Crystal Detection
Advanced imaging technologies are critical for accurate crystal identification, especially in high-throughput environments where thousands of images must be analyzed. Each modality offers specific advantages for different crystallization scenarios.
Visible Light Imaging: The standard technique captures images using the visible light spectrum (400-700 nm wavelength). While suitable for analyzing large protein crystals, it cannot distinguish between protein and salt crystals, presenting a significant limitation for automated analysis [8].
Ultraviolet (UV) Imaging: This label-free imaging modality illuminates protein drops with UV light, detecting fluorescence from aromatic amino acids like tryptophan to distinguish protein crystals from salt. However, it may yield false-positive results with phase separation and protein aggregation [8] [33].
Multi-Fluorescence Imaging (MFI): A powerful technique for imaging crystals of fluorescently labeled proteins using the trace fluorescent labeling (TFL) approach. MFI efficiently distinguishes protein crystals from salts, as well as crystals of single proteins from complexes, providing exceptional specificity in complex mixtures [8].
Second Order Nonlinear Imaging of Chiral Crystals (SONICC): This technology combines Second Harmonic Generation (SHG) with Ultraviolet Two-Photon Excited Fluorescence (UV-TPEF) to image protein crystals. It easily detects microcrystals (<1 μm) and crystals obscured in birefringent LCP or buried under aggregates, offering the highest sensitivity for challenging detection scenarios [8] [33].
Specialized Crystallization Methods
Automated systems support various protein crystallization techniques, each with distinct advantages for different protein types and experimental goals. The choice of method depends on multiple factors, including protein type, available volume, and desired outcomes [8].
Table 4: Comparison of Protein Crystallization Techniques
| Method | Amount of Protein | Automation | Seeding | Harvesting | Best Suitability |
|---|---|---|---|---|---|
| Sitting Drop [8] | Small | Possible | Possible | Easy | Initial screening |
| Hanging Drop [8] | Small to Large | Possible | Possible | Easy | Crystallization optimization using high surface tension reagents |
| Micro-Batch [8] | Small | Not Possible | Not Possible | Difficult | For proteins and reagents having minimal interactions with oil |
| Micro-Dialysis [8] | Large | Not Possible | Not Possible | Easy | For developing large crystals |
| Free-Interface Diffusion [8] | Very Small | Difficult | Possible | Difficult | For diffraction-quality crystals |
| Lipidic Cubic Phase [8] | Small to Large | Possible | Possible | Difficult | For high-quality crystals of membrane proteins |
Automation has fundamentally transformed protein crystallization from a manual, artisanal process to a systematic, high-throughput science. The integrated ecosystem of liquid handlers, crystallization robots, and automated storage imagers has dramatically improved the efficiency, reproducibility, and success rates of crystallization experiments while conserving precious protein samples [8] [30]. This technological evolution enables researchers to comprehensively explore crystallization parameter space by testing multiple protein constructs against hundreds of conditions, significantly accelerating structural biology research and drug discovery pipelines.
Future developments in automation continue to address remaining bottlenecks. The integration of AI-based autoscoring models like Sherlock with crystallization management software represents a significant advancement in handling the extensive image datasets generated by high-throughput systems [8]. As these algorithms continually improve through user feedback and machine learning, their accuracy in distinguishing true crystals from precipitate and salt will further reduce researcher workload and decrease analysis time. The ongoing refinement of nanoliter dispensing technologies, advanced imaging modalities, and integrated software solutions promises to make structural biology increasingly accessible, pushing the boundaries of our understanding of biological macromolecules and facilitating the development of novel therapeutics.
In high-throughput protein crystallization pipelines, accurately distinguishing protein crystals from salt crystals remains a significant challenge, with manual inspection leading to false negative rates as high as 20% [37]. Advanced imaging modalities have emerged as critical tools to overcome this bottleneck, enabling researchers to confidently identify hits and accelerate structural biology and drug discovery efforts. This application note details the operational principles, protocols, and practical implementation of three key technologies: Ultraviolet (UV) imaging, Multi-Fluorescence Imaging (MFI), and Second Order Nonlinear Imaging of Chiral Crystals (SONICC). By integrating these techniques into high-throughput workflows, researchers can significantly improve the efficiency and success of crystallization screening, ultimately advancing structure-based drug design.
Technical Comparison of Imaging Modalities
The following table summarizes the key characteristics of UV, MFI, and SONICC imaging technologies to guide appropriate selection and application.
Table 1: Comparative analysis of advanced imaging modalities for protein crystallization
| Feature | UV Imaging | Multi-Fluorescence Imaging (MFI) | SONICC |
|---|---|---|---|
| Fundamental Principle | Tryptophan fluorescence under ~295 nm excitation [37] | Fluorescence from dye-labeled proteins [38] | Second Harmonic Generation (SHG) from chiral crystals [39] |
| Detection Capability | Protein crystals containing tryptophan | Protein crystals (even tryptophan-free via labeling) [38] | Protein microcrystals (<1 μm) [39] |
| Key Strength | Label-free detection | Distinguishes protein-protein complexes [38] | Detects crystals in turbid media (e.g., LCP) [40] |
| Primary Limitation | Weak signal for tryptophan-free proteins; some salt interference [37] | Requires protein labeling with dyes [8] | Cannot distinguish between different chiral protein crystals [39] |
| Typical Imaging Time | ~10 minutes per 96-well plate [38] | ~5 minutes per 96-well plate (visible fluorescence) [38] | Varies by system and plate density |
| Sample Impact | Potential photo-damage with prolonged exposure [40] | Minimal known sample damage [38] | Non-destructive [39] |
Experimental Protocols & Methodologies
Protocol for UV Imaging
UV imaging leverages the intrinsic fluorescence of tryptophan residues in proteins when excited with UV light at approximately 295 nm [37]. The following protocol ensures optimal results:
- Sample Preparation: Crystallization trials should be set up in UV-transparent plates and sealed with low-UV-absorbing seals to allow maximum transmission of excitation and emission light [37].
- Image Acquisition: Program an automated UV imager (e.g., Rock Imager series) to illuminate drops with UV light at 295 ± 5 nm. A typical exposure time is 1200 milliseconds [37].
- Image Analysis: In the resulting images, protein crystals will typically fluoresce due to their tryptophan content, while salt crystals remain dark. Be aware that certain salts (e.g., primuline yellow) can be fluorescently active and create false positives, while proteins with very low tryptophan content may yield false negatives [37].
Protocol for Multi-Fluorescence Imaging
MFI requires pre-labeling proteins with fluorescent dyes but provides high-contrast images and can differentiate protein complexes [38]. The labeling and imaging protocol is as follows:
- Protein Labeling:
- Prepare a 5 mM stock solution of a succinimidyl ester dye (e.g., Fluorescein or Texas Red) in DMSO.
- Add the appropriate amount of dye to the protein solution to achieve a 0.1% labeling ratio of lysine residues, assuming 1:1 stoichiometric labeling efficiency.
- Incubate for 5 minutes; at this point, approximately 90% of the dye is bound. Purification is typically not required, and the labeled sample remains stable for over 120 days [38].
- Crystallization Setup: Set up crystallization trials using the labeled protein via standard methods (e.g., sitting or hanging drop vapor diffusion).
- Image Acquisition: Image the drops using an MFI-capable imager (e.g., Rock Imager 2 or 360) at the excitation/emission wavelengths specific to the chosen dyes. For studying complexes, label individual proteins or subunits with different dyes [38].
- Analysis: Protein crystals will fluoresce at the specific wavelength of the bound dye. For protein-protein complexes where two different dyes were used, crystals fluorescing at both wavelengths indicate a complex, while fluorescence at only one wavelength indicates a crystal of a single protein [38].
Protocol for SONICC Imaging
SONICC combines Second Harmonic Generation (SHG) and Ultraviolet Two-Photon Excited Fluorescence (UV-TPEF) to detect submicron crystals and crystals obscured in precipitate or lipidic cubic phase (LCP) [40] [39].
- Sample Preparation: Crystallization trials can be set up using standard methods, including LCP and microbatch. No labeling is required.
- Image Acquisition:
- SHG Mode: Use to identify chiral crystalline structures (a property of protein crystals). This produces high-contrast images where protein crystals appear bright white against a black background, even in turbid conditions [39].
- UV-TPEF Mode: Use in conjunction with SHG to confirm the protein nature of the crystals by detecting intrinsic fluorescence, helping distinguish protein crystals from other chiral crystals like sugar [40] [39].
- Analysis: Identify hits by locating bright signals in the SHG channel. Co-localization with UV-TPEF signal strongly confirms a protein crystal.
Workflow Integration and Data Analysis
The integration of UV, MFI, and SONICC into a high-throughput protein crystallization workflow significantly enhances hit identification and validation. The following diagram illustrates the synergistic relationship between these modalities and the critical decision points in the experimental process.
Automated image analysis is crucial for managing the large datasets generated by high-throughput workflows. The integration of AI-based autoscoring models, such as the Sherlock model integrated with Rock Maker software, can streamline the initial analysis of crystallization images [8]. These tools help researchers prioritize conditions for further inspection using the advanced modalities outlined in this document.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of these imaging technologies requires specific reagents and equipment. The following table details the key components for establishing these workflows.
Table 2: Essential research reagents and solutions for advanced imaging workflows
| Item | Function/Application | Examples/Specifications |
|---|---|---|
| UV-Transparent Plates/Seals | Allows transmission of UV light for excitation and emission detection in UV imaging [37]. | Plates and seals made from low-UV-absorbing materials (e.g., Cyclo-olefin polymer). |
| Amine-Reactive Dyes | Fluorescent labels for proteins in MFI; bind to lysine residues [38]. | Succinimidyl ester dyes (e.g., Fluorescein, Texas Red). Prepare as 5 mM stock in DMSO. |
| Automated Imaging Systems | Integrated platforms for high-throughput, automated image acquisition. | Rock Imager series (configurable with UV, MFI, SONICC) [38] [8] [39]. |
| Crystallization Screen Reagents | Sparse-matrix and statistical screening cocktails to sample crystallization chemical space [2]. | JCSG+ screen, various commercial and in-house screens. |
| Liquid Handling Robots | Automated setup of crystallization trials for precision and reproducibility with sub-microliter volumes. | NT8 Drop Setter, Formulator Screen Builder [8]. |
| Laboratory Information Management System (LIMS) | Software to manage the entire crystallization workflow, from experimental setup to data analysis. | Rock Maker software [8]. |
The integration of UV, MFI, and SONICC imaging technologies provides a powerful, multi-faceted approach to overcoming one of the most persistent challenges in high-throughput protein crystallization. By understanding the specific strengths and applications of each modality—UV for label-free detection, MFI for complex analysis and low-tryptophan proteins, and SONICC for microcrystals and turbid matrices—researchers can design more robust and successful workflows. As the protein crystallization market continues to grow, driven by demand in biopharmaceuticals and personalized medicine [9], the adoption of these advanced imaging techniques will be instrumental in accelerating drug discovery and structural biology research.
Specialized Approaches for Membrane Proteins and Large Complexes
Membrane proteins and large macromolecular complexes represent a significant frontier in structural biology, with profound implications for understanding cellular function and enabling structure-based drug discovery. Despite comprising approximately 30% of all proteins in living organisms and representing over 50% of current drug targets, membrane proteins constitute only about 1.5% of the structures in the Protein Data Bank [41]. This disparity highlights the unique challenges associated with their structural characterization. Unlike their soluble counterparts, membrane proteins require extraction from their native lipid environments using detergents and stabilization in solution for crystallization trials [42]. The entity being crystallized is not merely the protein itself, but rather the protein-detergent complex, introducing additional variables that complicate the crystallization process [42].
Large macromolecular complexes present their own distinct challenges, including structural heterogeneity, conformational flexibility, and difficulties in producing sufficient quantities of homogeneous sample. For both membrane proteins and large complexes, traditional crystallization approaches developed for soluble proteins often prove inadequate, necessitating specialized methodologies tailored to their unique properties. This application note details advanced strategies and protocols developed to address these challenges, enabling researchers to overcome traditional bottlenecks in structural determination of these biologically significant targets.
Key Research Reagent Solutions for Membrane Protein Crystallization
The following table summarizes essential reagents and materials specifically valuable for membrane protein crystallization workflows:
Table 1: Key Research Reagent Solutions for Membrane Protein Crystallization
| Reagent/Material | Function/Application | Examples/Notes |
|---|---|---|
| Detergents | Solubilize and stabilize membrane proteins in aqueous solution | n-Dodecyl-β-D-maltopyranoside (DDM), n-Decyl-β-D-maltopyranoside (DM), n-Octyl-β-D-glucopyranoside (OG) [42] |
| Specialized Crystallization Screens | Rationally designed initial condition screening | MemGold, MemGold2 (for alpha-helical membrane proteins) [42] |
| Additive Screens | Optimization of initial crystal hits | MemAdvantage (contains additives specifically beneficial for membrane proteins) [42] |
| Lipidic Cubic Phase (LCP) Materials | Creating membrane-mimetic environment for crystallization | Monoolein lipid matrices [43] [41] |
| Selenium-labeled Lipids | Experimental phasing for in meso structures | Se-MAG (enables SAD/MAD phasing for LCP-grown crystals) [41] |
| Protein Engineering Tags | Enhancing protein stability and crystallization | T4 lysozyme, BRIL fusions (introduce crystallization scaffolds) [42] |
Quantitative Analysis of Successful Membrane Protein Crystallization Conditions
Analysis of successful membrane protein crystallization experiments reveals distinct trends in detergent selection and precipitant usage that differ markedly from conditions optimal for soluble proteins. The table below summarizes key statistical findings from empirical data:
Table 2: Statistical Analysis of Successful Membrane Protein Crystallization Conditions from Empirical Data
| Parameter | Trend/Optimal Condition | Statistical Basis |
|---|---|---|
| Most Successful Detergent Class | Alkyl Maltopyranosides | Account for 50% of all reported structures [42] |
| Highest Resolution Detergent | Alkyl Glucopyranosides (particularly OG) | Mean resolution of 2.5 Å; highest resolution structure at 0.88 Å [42] |
| Second Highest Resolution Detergent Class | Amine Oxides (e.g., LDAO) | Mean resolution of 2.66 Å [42] |
| Most Successful Alkyl Maltopyranoside | n-Dodecyl-β-D-maltopyranoside (DDM) | Most frequently used successful detergent in class [42] |
| Primary Precipitants | Polyethylene Glycol (PEG) variants | More successful than high-salt conditions commonly used for soluble proteins [42] |
| Functional Family Success Rate | Channels and Transporters | Highest number of determined structures (149 and 157 respectively) [42] |
Specialized Experimental Protocols
Protocol: High-Throughput Crystallization Screening for Membrane Proteins Using Lipidic Cubic Phase (LCP) Method
Principle: The LCP method (also called in meso) creates a membrane-mimetic environment that maintains membrane proteins in a more native lipid bilayer context, often leading to better-ordered crystals [43] [41]. This protocol adapts the method for high-throughput implementation.
Materials:
- Purified membrane protein in appropriate detergent (≥ 5 mg/mL)
- Monoolein or similar lipid
- LCP-capable crystallization plates (e.g., 96-well glass sandwich plates)
- Oryx 4 crystallization robot or similar system with LCP module [43]
- Pre-formulated crystallization screens (MemGold2, MemStart)
- Syringe mixer or similar device for LCP formation
Procedure:
- LCP Formation: Mix purified membrane protein with molten monoolein (typically 60:40, lipid:protein ratio by volume) using a syringe mixer until a clear, viscous cubic phase is obtained. This is evidenced by uniform consistency and optical clarity.
- Plate Setup: Using the LCP module of the crystallization robot, dispense 50-100 nL LCP boluses onto the plate substrate followed by 800-1000 nL of precipitant solution from crystallization screens.
- Sealing: Immediately seal the plate with a transparent cover seal to prevent evaporation.
- Incubation: Incubate plates at controlled temperatures (typically 20°C and 4°C) for crystallization trials.
- Monitoring: Image plates regularly using automated imaging systems (e.g., Rock Imager series) with capabilities for UV imaging or SONICC to detect microcrystals [8].
Technical Notes: Protein concentration and lipid-to-protein ratio are critical parameters that may require optimization. LCP crystallization often produces very small crystals, requiring advanced imaging techniques for detection. The protocol can be modified to include selenium-labeled lipids (Se-MAG) for direct experimental phasing by replacing half of the monoolein with Se-MAG [41].
Protocol: Detergent Optimization for Membrane Protein Crystallization
Principle: Identifying the optimal detergent and detergent combination is often the most critical step in membrane protein crystallization. This systematic approach screens for detergents that enhance stability and promote crystal formation.
Materials:
- Purified membrane protein in initial extraction detergent (e.g., DDM)
- Detergent exchange columns (e.g., size exclusion)
- Range of detergents including alkyl maltopyranosides, glucopyranosides, and amine oxides
- Thermofluor stability assay components or similar stability assessment kit
- Crystallization robots (e.g., NT8 Drop Setter) for high-throughput setup [8]
Procedure:
- Detergent Screening: Set up small-scale (50-100 μL) detergent exchanges into 8-12 different detergents spanning various classes (DDM, DM, OG, LDAO, etc.).
- Stability Assessment: Evaluate protein stability in each detergent using thermal shift assays, monitoring both melting temperature (Tm) and aggregation onset.
- Crystallization Trials: Set up initial crystallization screens (MemGold1 & 2) with the 3-4 most stable detergent conditions using vapor diffusion in sitting drop format with 100 nL drop size.
- Additive Screening: For conditions showing promising hits (microcrystals, phase separation), set up additive screens using MemAdvantage or similar, incorporating 2-5% additive concentrations.
- Detergent Mixing: For proteins that remain unstable in single detergents, systematically test mixed detergent systems, beginning with combinations of maltosides and glucosides.
Technical Notes: Shorter-chain detergents (e.g., OG vs DDM) often produce higher-resolution crystals but may compromise stability [42]. Always assess protein monodispersity after detergent exchange using analytical size exclusion chromatography. The use of detergent cocktails (mixed detergents) has increasingly been successful for challenging targets [42].
Workflow Visualization and Decision Pathways
The following diagram illustrates the integrated experimental workflow for membrane protein crystallization, incorporating both traditional and specialized approaches:
Diagram 1: Membrane Protein Crystallization Workflow
Advanced and Emerging Techniques
Computational Protein Design for Soluble Membrane Protein Analogues
Recent breakthroughs in deep learning-based protein design have enabled the creation of soluble analogues of integral membrane protein folds, potentially revolutionizing drug discovery approaches. By inverting AlphaFold2 networks combined with ProteinMPNN sequence optimization, researchers have successfully designed stable soluble versions of previously inaccessible membrane protein topologies including GPCRs, claudins, and rhomboid proteases [44]. This approach effectively expands the soluble fold space to include previously membrane-restricted topologies, enabling structural and functional studies without detergent requirements. The method has demonstrated remarkable experimental success rates, with designed proteins showing high thermal stability and accurate structural formation [44].
Advanced Imaging and Automation Technologies
Modern membrane protein crystallography increasingly relies on advanced imaging technologies to detect and characterize often microscopic crystals. Second Order Non-linear Imaging of Chiral Crystals (SONICC) provides exceptional sensitivity for detecting microcrystals (<1 μm) that would be invisible with traditional brightfield microscopy, particularly those obscured in birefringent LCP matrices or buried under aggregates [8]. When combined with Ultraviolet Two-Photon Excited Fluorescence (UV-TPEF), this approach can reliably distinguish protein crystals from salt crystals. Furthermore, fully integrated robotic systems such as the Formulatrix platform (including Rock Maker software, Formulator screen builder, NT8 drop setter, and Rock Imagers) enable complete automation of the crystallization workflow from screen preparation to image analysis, dramatically increasing throughput and reproducibility while minimizing human error and sample consumption [8].
The specialized approaches outlined in this application note provide researchers with a comprehensive toolkit for addressing the unique challenges presented by membrane proteins and large complexes. The integration of rationally designed screens like MemGold, membrane-mimetic environments such as LCP, targeted additive strategies, and advanced detection methods significantly increases the probability of successful structure determination. Furthermore, emerging technologies in computational protein design and automated workflow integration promise to accelerate progress in this critical field. By implementing these specialized protocols and leveraging the statistical trends revealed through empirical analysis of successful structures, researchers can systematically overcome traditional bottlenecks and advance our understanding of these biologically essential but structurally challenging targets.
From Hits to High-Quality Crystals: Advanced Optimization and AI-Driven Troubleshooting
Within high-throughput protein crystallization pipelines, the initial identification of "lead conditions" or "hits" from a broad screen is a crucial first step. However, these initial conditions seldom yield crystals of diffraction-quality without systematic refinement [6] [45]. This application note details a structured protocol for the systematic optimization of these lead conditions, focusing on the interdependent variables of precipitant concentration, pH, and temperature. The objective is to guide researchers from microcrystalline hits or poorly diffracting crystals to well-ordered, single crystals suitable for high-resolution X-ray data collection, thereby enhancing the efficiency of structural genomics and drug development programs [46] [45].
Background and Principles
Protein crystallization is a process of phase separation driven by the sample's traversal of a phase diagram from an undersaturated state into a metastable, supersaturated zone where nucleation and crystal growth can occur [47]. Initial screening identifies conditions that place the protein in this productive zone. Optimization aims to fine-tune these conditions to first control nucleation and then promote the steady growth of large, ordered crystals [45].
The process is inherently multidimensional. Key chemical parameters include the type and concentration of precipitating agents, the pH of the solution, and the ionic strength. Physical parameters such as temperature and sample volume also exert significant influence [45]. A fundamental challenge is that these parameters are often interdependent; a change in temperature can affect pH, while a change in pH can alter protein solubility and precipitant efficacy [45]. Therefore, a systematic, rather than a univariate, approach is recommended.
Key Optimization Parameters and Strategies
Precipitant Fine-Tuning
Precipitants work by reducing the solubility of the protein, driving the system toward supersaturation. They can operate through different mechanisms, such as excluded volume (e.g., polyethylene glycols) or "salting-out" (e.g., ammonium sulfate) [47].
- Systematic Concentration Variation: The most straightforward strategy is to vary the concentration of the primary precipitant around the hit condition in fine increments. For a hit condition containing 20% PEG 3350, one might set up a grid testing concentrations from 15% to 25% in 1-2% increments [45].
- Precipitant Synergy: Research has demonstrated that combining mechanistically distinct precipitants (e.g., a salt and an organic solvent) can significantly enhance the probability of crystallization and crystal quality by creating unique lattice interactions [48]. Consider designing conditions that blend two precipitants from different classes found in initial hits.
pH Optimization
The pH of the crystallization condition profoundly affects the ionization state of surface residues, influencing the protein's solubility and its ability to form specific, ordered crystal contacts [47] [49]. Proteins frequently crystallize within 1-2 pH units of their isoelectric point (pI) [47].
A high-throughput pH optimization involves using a fixed set of buffers covering a broad pH range (e.g., pH 4.0 to 9.0) while keeping other components constant [49]. This can be efficiently executed using automated liquid handlers to dispense a matrix of conditions where the pH is the primary variable.
Temperature Control
Temperature is a powerful but often underexploited parameter. It directly affects solubility, nucleation kinetics, and crystal growth rates.
- Incubation Temperature: Screening at different constant temperatures (e.g., 4°C, 12°C, 20°C) can identify the optimal thermal environment for crystal growth [50].
- Controlled Cooling: For crystals grown by batch or microbatch methods, a controlled reduction in temperature can slowly increase supersaturation, promoting steady crystal growth. Cooling-rate screening tools have been developed to identify the ideal cooling profile, which can prevent excessive nucleation and lead to larger, higher-quality crystals [51].
Table 1: Summary of Key Optimization Parameters and Their Effects
| Parameter | Experimental Range | Primary Effect | Optimization Goal |
|---|---|---|---|
| Precipitant Concentration | ± 10-30% of hit condition | Modifies supersaturation level | To find a balance between nucleation and growth |
| pH | Hit pH ± 1.5 units | Alters protein surface charge and solubility | To identify the point of optimal crystal packing |
| Temperature | 4°C, 12°C, 20°C, or controlled cooling | Impacts kinetics and solubility | To control nucleation density and growth rate |
| Additives | Ligands, ions, small molecules | Stabilizes specific conformations; mediates contacts | To improve crystal order and diffraction quality |
Integrated Experimental Protocols
Protocol 1: Iterative Screen Optimization (ISO)
Iterative Screen Optimization is a highly automated, data-driven method for refining crystallization conditions [46].
- Initial Screening: Set up a first-pass 96-well crystallization screen (e.g., a custom "Sweet16" screen or commercial equivalent) using a sitting-drop vapor diffusion or microbatch-under-oil method.
- Scoring and Analysis: After incubation, manually inspect each drop and assign a qualitative score (e.g., 0 for clear, 1 for precipitate, 2 for microcrystals, 3 for single crystals).
- Algorithmic Reformulation: Feed the scores into an optimization algorithm. This software calculates new precipitant concentrations for each of the 96 conditions, increasing concentrations for clear drops and decreasing them for heavily precipitated drops, effectively "tuning" the entire screen.
- Iteration: Use an automated liquid handler to set up the newly formulated screen. Repeat the scoring and reformulation process for several rounds. With each iteration, the screen becomes more tailored to the target protein, systematically exploring the precipitant concentration parameter space [46].
Protocol 2: High-Throughput pH and Precipitant Grid Screening
This protocol uses automation to create a fine-sampling 2D grid around a promising lead condition.
- Stock Solution Preparation: Prepare a concentrated stock solution of the mother liquor from the lead condition, omitting the buffer.
- Buffer Array: Obtain a set of buffering solutions covering a targeted pH range (e.g., Sodium Acetate for pH 4.0-5.5, Bis-Tris for pH 5.5-7.0, HEPES for pH 7.0-8.0, Tris for pH 8.0-9.0).
- Liquid Handling Setup: Program an automated liquid handler to:
- Dispense a constant volume of each buffer into wells of a 96-well crystallization plate.
- Add the mother liquor stock to each well, creating a series of solutions with identical precipitant and additive composition but varying pH.
- For a 2D grid, set up parallel rows where the precipitant concentration is also varied.
- Crystallization Trial: Dispense the protein solution into each drop to initiate the crystallization trial. This protocol efficiently decouples pH from other variables for direct analysis [49].
The following diagram illustrates the logical decision-making process for navigating the optimization workflow, from initial hit analysis to the selection of specific protocols.
The Scientist's Toolkit: Essential Reagents and Materials
Successful optimization relies on a suite of specialized reagents and equipment designed to enable precise, high-throughput experimentation.
Table 2: Key Research Reagent Solutions for Optimization
| Item Name | Function/Description | Application in Optimization |
|---|---|---|
| Polyethylene Glycols (PEGs) | A family of polymers that act as excluded-volume precipitants. Available in a range of molecular weights (400 - 8000 Da). | The most common precipitant class; concentration and molecular weight are key variables to test. [46] [45] |
| Ammonium Sulfate | A classic "salting-out" precipitant. | Used to explore a different precipitant mechanism than PEG; concentration is critical. [47] |
| 2-methyl-2,4-pentanediol (MPD) | An organic solvent precipitant and common additive. | Binds to hydrophobic protein regions, affecting the hydration shell; can be used as a co-precipitant or cryoprotectant. [47] [46] |
| Buffers (e.g., Bis-Tris, HEPES) | Solutions to control the pH of the crystallization condition. | Essential for pH optimization grids; should be selected for stability over the desired pH range. [46] [49] |
| Tris(2-carboxyethyl)phosphine (TCEP) | A reducing agent with a long solution half-life across a wide pH range. | Maintains protein stability by preventing cysteine oxidation during long crystallization trials, crucial for reproducible results. [47] |
| Crystallization Plates (96-/384-well) | Plastic plates with wells for setting up nanoliter- to microliter-volume trials. | The standard platform for high-throughput vapor diffusion or microbatch experiments. [52] |
| Automated Liquid Handler | Robotic system for dispensing nanoliter volumes of protein and screen solutions. | Enables the setup of complex, high-density optimization grids with high reproducibility and minimal protein consumption. [46] [52] |
Discussion
The optimization strategies outlined here transform the art of crystal growth into a more systematic, data-driven science. The core challenge remains the interdependency of parameters; a change in pH can shift the optimal precipitant concentration, which in turn may be sensitive to temperature [45]. Therefore, after initial univariate screens, it is often necessary to perform small multivariate experiments to capture these interactions.
The choice of initial hits to optimize is critical. Dense microcrystalline showers, fractal structures, or thin plates can be difficult to improve. In contrast, small three-dimensional crystals or single crystals with poor diffraction are often the most promising candidates for optimization [45]. Furthermore, integrating computational prediction tools early in the process can enhance success. Analyzing targets with AlphaFold to remove flexible regions or using "crystallizability" scores for construct design can yield more stable, crystallization-prone protein samples from the outset [47] [50].
For proteins that remain recalcitrant, advanced techniques such as seeding—using microcrystals from a hit to nucleate growth in optimized, less supersaturated conditions—or the use of crystallization chaperones like engineered antibody fragments can provide a path forward [47] [6].
Systematic optimization of lead conditions is a non-negotiable step in high-throughput structural biology. By methodically fine-tuning precipitant concentration, pH, and temperature, researchers can significantly increase their odds of transforming initial promising signs into diffraction-quality crystals. The protocols for Iterative Screen Optimization and high-throughput grid screening, supported by automated liquid handling and a well-stocked toolkit of reagents, provide a robust framework for this endeavor. As structural genomics continues to tackle more challenging targets, such as membrane proteins and large complexes, these disciplined optimization practices will become increasingly vital for accelerating drug discovery and deepening our molecular understanding of biological processes.
Within high-throughput structural biology pipelines, obtaining diffraction-quality crystals remains a significant bottleneck. Initial crystallization screens often yield microcrystals, phase separation, or precipitate, requiring systematic optimization to produce crystals suitable for X-ray diffraction analysis [2] [6]. This Application Note details two powerful, complementary techniques—seeding and additive screening—that leverage initial crystallization "hits" to improve crystal size, morphology, and diffraction quality. These methods are essential for advancing structural studies of therapeutic targets and complex macromolecular assemblies.
Seeding Techniques
Seeding is a fundamental optimization strategy that separates the processes of crystal nucleation and growth. By introducing pre-formed crystalline material (seeds) into new crystallization experiments, researchers can bypass the stochastic nucleation barrier, promote growth at lower supersaturation levels, and systematically improve crystal quality [53] [54].
Comparison of Seeding Methods
The table below summarizes the key characteristics, applications, and requirements of major seeding techniques.
Table 1: Comparison of Protein Crystallization Seeding Methods
| Method | Principle | Best For | Throughput | Key Equipment |
|---|---|---|---|---|
| Streak Seeding [55] [54] | Transfer of microseeds via fibrous tool (e.g., cat whisker, horse hair) | Rapidly testing a few new conditions | Low | Microscopic tool, crystallization plates |
| Seed Bead [54] | Mechanical crushing of crystals with a bead to create a "seed stock" | Creating a reusable seed resource for multiple experiments | Medium | Seed bead (e.g., Hampton Research kit), vortexer |
| Matrix Microseeding [56] [55] | Titrating diluted seed stock across a wide range of chemical conditions | High-throughput optimization and rescuing poorly diffracting crystals | High | Liquid handling robot, 96-well plates, seed stock |
| Macroseeding [53] | Transfer and washing of a single crystal to a new drop for further growth | Significantly increasing the size of a single, well-ordered crystal | Low | Micro-tools, stereomicroscope |
Detailed Protocols
Seed Bead and Seed Stock Preparation
This protocol creates a reusable seed stock from existing crystals [54].
- Harvest Crystals: Using a micropipette, transfer 5-10 µL of the mother liquor containing the microcrystals or crystalline precipitate into a microcentrifuge tube.
- Add Seed Bead: Place a single seed bead (e.g., from a Hampton Research Seed Bead kit) into the tube.
- Crush Crystals: Vortex the tube vigorously for 10-30 seconds. The goal is to create a homogeneous suspension of microscopic crystal fragments.
- Prepare Dilutions: Centrifuge the seed stock for 30 seconds at low speed (1-2 x g) to pellet large debris. Serially dilute the supernatant (e.g., 1:10, 1:100, 1:1000) in a solution matching the mother liquor or a stabilizing buffer.
- Storage: Keep the seed stock and its dilutions on ice to prevent premature dissolution of seeds.
Matrix Microseeding Protocol
This high-throughput protocol systematically screens for improved growth conditions [56] [55].
- Prepare Crystallization Plates: Dispense reservoir solutions (e.g., commercial screens or optimization grids) into 96-well plates.
- Formulate Drops: Using a liquid handling robot (e.g., Mosquito, Oryx), mix the following in the experimental well:
- 150 nL of purified protein sample.
- 200 nL of reservoir solution.
- 50 nL of diluted seed stock.
- Incubate and Monitor: Seal the plates and incubate at the appropriate temperature. Monitor for crystal growth regularly using an automated imaging system.
Robotic In-Situ Proteolysis Seeding
Proteases can be added as "seeds" to optimize crystallization of difficult targets [56].
- Protease Stock: Prepare a stock solution of protease (e.g., trypsin) at 0.1 mg/mL in a compatible buffer (e.g., 10 mM Tris-HCl pH 8.0).
- Robot Setup: Program a crystallization robot (e.g., Oryx) to dispense crystallization drops with a variable "seed factor" for the protease volume.
- Drop Setup: A typical drop might consist of:
- 0.3 µL protein solution (8-10 mg/mL)
- 0.25 µL reservoir solution
- 0.01-0.05 µL protease stock (varied in 0.01 µL steps across different drops)
- Optimization: The optimal protease concentration is identified by comparing crystal number, size, and diffraction quality across the different drops.
Additive Screening
Additive screening involves introducing small molecule compounds or reagents into crystallization experiments to fine-tune molecular interactions and improve crystal order.
Additive Classes and Functions
Table 2: Common Additive Classes and Their Proposed Mechanisms of Action
| Additive Class | Example Compounds | Proposed Mechanism | Impact on Crystals |
|---|---|---|---|
| Ions & Salts [55] | NiSO₄, CaCl₂, ZnCl₂ | Modulate surface charge and mediate specific crystal contacts | Can dramatically change crystal habit (e.g., from needles to plates) |
| Reducing Agents | TCEP, DTT | Stabilize cysteine residues and prevent disulfide bond heterogeneity | Improves reproducibility and diffraction limit |
| Substrates/Inhibitors [55] | Co-factors, substrate analogs | Stabilize a specific, homogeneous protein conformation | Often essential for obtaining crystals of ligand-binding proteins |
| Ionic Liquids [52] | Imidazolium-based salts | Alter water activity and solvent properties | Can reduce nucleation density and improve crystal size |
High-Throughput Additive Screening Protocol
- Additive Library: Obtain a commercial additive screen (e.g., from Hampton Research) or prepare a custom library of potential additives.
- Plate Setup: Use a liquid handler to dispense reservoir solution into 96-well plates.
- Additive Incorporation: Add a small, defined volume (e.g., 50 nL) of each additive solution directly to the crystallization drop or to the reservoir solution.
- Experimental Setup: Mix the protein solution with the additive-containing reservoir solution.
- Analysis: Use automated imaging to identify additives that lead to crystal formation or improved crystal morphology. Second-order nonlinear imaging of chiral crystals (SONICC) can be particularly useful for detecting microcrystals obscured by precipitate [52] [8].
Workflow Integration and Data Management
Integrating seeding and additive screening into a high-throughput pipeline requires careful planning and data management. The diagram below illustrates a recommended automated workflow.
The Scientist's Toolkit
Successful implementation of these advanced techniques relies on specific reagents and instrumentation.
Table 3: Essential Research Reagent Solutions and Materials
| Item | Function/Application | Example Products/Suppliers |
|---|---|---|
| Seed Beads | Mechanical crushing of crystals to create microseed stock. | Hampton Research Seed Bead Kits |
| Additive Screens | Systematic introduction of small molecules to optimize crystal contacts and stability. | Hampton Research Additive Screen, Molecular Dimensions Classic Additive Screen |
| Proteases | Limited proteolysis to remove flexible termini/loops, improving protein homogeneity for crystallization. | Trypsin, α-Chymotrypsin (e.g., Sigma-Aldrich) |
| Liquid Handler | Automated, nanoliter-dispensing for high-throughput and reproducible screen setup. | TTP LabTech Mosquito, Formulatrix NT8, Douglas Instruments Oryx |
| Automated Imager | Regular, non-invasive monitoring of crystal growth over time. | Formulatrix Rock Imager series [8] |
| AI Scoring Software | Automated analysis of large image datasets to identify crystal hits. | MARCO Polo, Sherlock [52] [8] |
Seeding and additive screening are powerful, synergistic techniques essential for transforming initial crystallization hits into diffraction-quality crystals. The protocols outlined herein, when integrated into a high-throughput workflow supported by automation and AI-assisted analysis, provide a robust framework for accelerating structural biology research and structure-based drug discovery.
Leveraging AI and Machine Learning for Intelligent Condition Prediction and Autoscoring
The process of protein crystallization has long been a critical bottleneck in structural biology, limiting the pace of drug discovery and functional analysis. High-throughput crystallization screening has emerged as a technological solution, allowing researchers to test thousands of biochemical conditions in parallel [2]. However, this approach generates enormous datasets of crystallization images and outcomes that present new challenges in analysis and interpretation. Artificial intelligence (AI) and machine learning (ML) are now revolutionizing this field by introducing intelligent condition prediction and automated image scoring (autoscoring) capabilities [8]. These technologies are transforming protein crystallization from an empirical art to a data-driven science, enabling researchers to navigate complex chemical spaces more efficiently and extract meaningful insights from extensive experimental results. This application note details protocols and methodologies for implementing AI and ML in high-throughput protein crystallization workflows, with specific focus on condition prediction and autoscoring applications relevant to drug development professionals.
AI for Intelligent Crystallization Condition Prediction
Machine Learning Approaches for Co-crystal Formation
Machine learning algorithms have demonstrated significant potential in predicting successful co-crystal formation for pharmaceutical compounds. One innovative approach utilizes a link prediction algorithm that analyzes crystallographic data to identify promising co-formers [57].
Protocol: AI-Guided Co-crystal Screening
- Data Collection and Network Mapping: Extract all known co-crystal structures from the Cambridge Structural Database (CSD), which contains over one million crystal structures determined at Å resolution. Map the relationships between APIs and co-formers as an interconnected network, analogous to social network analysis [57].
- Algorithmic Analysis: Apply machine learning link prediction algorithms to identify missing connections between nodes (APIs and potential co-formers) for similar molecules. The algorithm analyzes the correlation network to predict which API-co-former pairs are most likely to form co-crystals [57].
- Experimental Validation: Test the top-ranked co-former predictions using high-throughput crystallization platforms. The Crystal16 instrument provides medium-throughput capabilities for co-crystal screening, allowing testing of one co-former in four solvents simultaneously with four concentrations each within a single day [57].
Table 1: Comparison of AI Approaches for Crystallization Condition Prediction
| AI Approach | Data Source | Application | Advantages | Limitations |
|---|---|---|---|---|
| Link Prediction Algorithm | Cambridge Structural Database (CSD) | Co-crystal formation prediction | Based on experimental crystallographic data; Reduces number of experiments required | Limited to compounds with structural analogs in database |
| Diffusion Generative Modeling | Materials Project database (>150,000 materials) | Atomic structure determination from powder diffraction | Solves previously intractable structures; Works with nanocrystals | Requires extensive training dataset; 67% accuracy on natural minerals |
| Autoscoring Integration (Sherlock) | Historical crystallization image datasets | Classification of crystallization outcomes | Continuously improved with user feedback; Integrates with existing workflows | Dependent on quality and diversity of training data |
Deep Learning for Structural Determination from Nanocrystals
Determining atomic structures from nanocrystalline materials has represented a century-old challenge in crystallography. Recent advances in deep learning architectures have enabled breakthroughs in this area through techniques adapted from AI-generated art programs [58].
Protocol: Nanocrystal Structure Determination with Crystalyze
- Model Training: Train a generative AI model on a dataset of 40,000 known atomic structures from the Materials Project database. Employ diffusion generative modeling, which begins with jumbled atomic positions and trains a deep neural network to connect these with associated X-ray diffraction patterns [58] [59].
- Structure Prediction: The model breaks the prediction process into subtasks: first determining the size and shape of the lattice "box," then predicting the arrangement of atoms within it. For each diffraction pattern, the model generates multiple possible structures (up to 100 guesses) [59].
- Validation Cycle: Test the predicted structures by feeding them into a model that simulates diffraction patterns. Compare the input pattern with the simulated output from the prediction; successful reconstructions show near-identical patterns [59].
- Structure Refinement: Submit AI-generated crystals through Rietveld refinement, a procedure that "jiggles" crystals into the closest optimal state based on the diffraction pattern [58].
Automated Workflows and AI-Driven Autoscoring
Integrated Automated Crystallization Platforms
Comprehensive automation solutions now encompass the entire protein crystallization workflow, from screen preparation to final imaging and analysis. These integrated systems provide the foundational infrastructure necessary for implementing AI and ML technologies effectively [8].
Table 2: Automated Protein Crystallization Workflow Components
| Workflow Stage | Technology | Key Features | Throughput Benefits |
|---|---|---|---|
| Screen Building | Formulator Microfluidic Dispenser | Dispenses up to 34 different ingredients with 96-nozzle chip; Volumes down to 200 nL | Prepares 100 μL, 3-ingredient grid across 96 wells in 2.7 minutes |
| Drop Setting | NT8 Drop Setter | Dispenses drops from 10 nL to 1.5 μL; Supports hanging/sitting drops, LCP, additives, seeding | Active humidification prevents evaporation; Reusable tips reduce costs |
| Imaging | Rock Imager Series | Various models with capacities from 1 to 1000 plates; Multiple imaging modalities (Visible, UV, MFI, SONICC) | Automated plate storage and retrieval with refrigeration |
| Data Management | Rock Maker Software | Integrates all crystallization products; Manages entire workflow including data analysis | Enables seamless AI autoscoring integration |
AI-Based Autoscoring Systems
The integration of AI-based autoscoring models has addressed one of the most time-consuming aspects of high-throughput crystallization: the analysis of extensive image datasets generated during experiments. These systems automatically classify crystallization outcomes, significantly reducing manual review time [8].
Protocol: Implementation of AI Autoscoring with Sherlock
- System Integration: Implement Sherlock, Formulatrix's proprietary autoscoring model, within the Rock Maker crystallization software environment. Ensure seamless data transfer between the imaging systems and analysis software [8].
- Continuous Model Enhancement: The autoscoring model undergoes continuous improvement based on feedback from users across the Rock Maker platform. This iterative learning process progressively enhances the model's performance and classification accuracy [8].
- Image Analysis and Classification: The AI model analyzes crystallization images using pattern recognition algorithms trained on thousands of classified examples. It distinguishes between clear drops, precipitation, microcrystals, and diffraction-quality crystals [8].
- Result Validation and Expert Review: While autoscoring handles the initial classification, the system flags ambiguous results for expert review, creating a hybrid human-AI workflow that maximizes both efficiency and accuracy [8].
Experimental Protocols for AI-Enhanced Crystallization
High-Throughput Screening with Integrated Autoscoring
This protocol details a complete workflow for high-throughput crystallization screening incorporating AI-based condition selection and autoscoring technologies.
Materials and Reagents
- Purified protein sample (>95% purity)
- Commercial sparse matrix screening solutions (e.g., Hampton Research, Molecular Dimensions)
- Formulator Screen Builder or equivalent liquid handling system
- NT8 Drop Setter or equivalent crystallization robot
- Rock Imager with UV or MFI capabilities
- Rock Maker software with Sherlock autoscoring integration
Procedure
- Protein Quality Control: Assess protein purity and concentration using SDS-PAGE and spectrophotometry. Confirm protein stability under screening conditions using dynamic light scattering or thermal shift assays.
- Condition Selection: Utilize AI-guided condition prediction to select initial screening parameters, prioritizing conditions with high success probability based on similar protein characteristics.
- Plate Setup: Program the liquid handling system to dispense screening solutions into 96-well crystallization plates. Use the drop setter to create sitting drop vapor diffusion experiments with 100-200 nL protein-to-reservoir solution ratios.
- Incubation: Seal plates and incubate at constant temperatures (4°C, 20°C) with monitoring via active humidification systems to prevent evaporation.
- Automated Imaging: Schedule regular imaging sessions (days 1, 3, 7, 14, 21, 30) using the automated imager with multiple modalities (brightfield, UV, MFI).
- AI Autoscoring: Enable Sherlock autoscoring to analyze all images as they are captured. The system automatically classifies outcomes and flags potential hits for review.
- Hit Identification and Validation: Review AI-classified hits manually to confirm results. Use second-order imaging techniques (SONICC) to distinguish protein crystals from salt when necessary.
Fragment Screening with CrystalDirect Technology
The CrystalDirect technology developed at EMBL Grenoble enables fully automated crystal harvesting and screening, particularly valuable for fragment-based drug discovery campaigns [60].
Materials and Reagents
- CrystalDirect harvester system
- Echo Labcyte acoustic dispensing system
- Pre-grown protein crystals
- Fragment library dissolved in DMSO
- Cryo-protectant solutions
Procedure
- Crystal Preparation: Grow protein crystals using optimized conditions identified through initial screening.
- Fragment Soaking: Use the acoustic dispensing system to transfer fragment solutions directly onto individual crystals. Optimize soaking times based on preliminary experiments.
- Automated Harvesting: Employ the CrystalDirect harvester to automatically cryo-cool and mount crystals for data collection. The system can process up to 400 crystals per operation cycle.
- Data Collection: Perform X-ray diffraction experiments at synchrotron beamlines, ideally integrated with the automated harvesting system.
- Data Processing: Use automated processing pipelines (e.g., CRIMS software) to quickly assess fragment binding and structural changes.
- AI-Enhanced Analysis: Implement ML algorithms to identify binding patterns across multiple fragments, prioritizing hits for further optimization.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagent Solutions for AI-Enhanced Crystallization
| Item | Function | Application Notes |
|---|---|---|
| Rock Maker Software | Laboratory Information Management System | Manages entire crystallization workflow; Integrates AI autoscoring; Provides data analysis tools |
| Formulator Screen Builder | Automated screen preparation | Dispenses nanoliter volumes of screening solutions; Accommodates all microplate types |
| NT8 Drop Setter | Crystallization robot | Sets up vapor diffusion experiments; Supports various methods (sitting/hanging drop, LCP) |
| Rock Imager Series | Automated imaging systems | Captures high-quality images of crystallization drops; Various modalities distinguish crystal types |
| Sherlock AI Model | Autoscoring of crystallization images | Classifies experimental outcomes; Continuously learns from user feedback |
| Crystal16 | High-throughput co-crystal screening | Provides medium-throughput platform; Measures light transmission during crystallization |
| CrystalDirect Harvester | Automated crystal processing | Harvests and cryo-cools crystals without manual intervention; Essential for fragment screening |
The integration of AI and machine learning technologies into high-throughput protein crystallization workflows represents a paradigm shift in structural biology research. These tools enable intelligent condition prediction that dramatically reduces experimental overhead and autoscoring systems that alleviate the analytical bottleneck of large-scale screening campaigns. For drug development professionals, these advancements translate to accelerated structure-based drug design, particularly for challenging targets like membrane proteins and macromolecular complexes. The protocols and methodologies detailed in this application note provide a framework for implementing these technologies effectively, with the potential to significantly enhance the efficiency and success rates of crystallization pipelines in both academic and industrial settings.
Visualizations
In high-throughput structural genomics pipelines, the production of diffraction-quality crystals remains a significant bottleneck, with only an estimated ~15% of purified proteins ultimately yielding a three-dimensional structure [61] [2]. The majority of experimental failures manifest as clear drops with no formation, promiscuous precipitate, or microcrystals that are unsuitable for X-ray diffraction. Success in structural biology depends on diagnosing the physical and chemical causes of these outcomes and implementing targeted salvage strategies. This application note details evidence-based protocols to address these common failures, leveraging methods such as chemical modification of protein surfaces, optimization of biochemical and physical parameters, and intelligent screening design to increase the success rate of crystallization campaigns.
Diagnostic Guide to Common Crystallization Outcomes
A critical first step is the accurate diagnosis of crystallization trial outcomes. The table below categorizes common results, their probable causes, and initial recommended actions.
Table 1: Diagnostic Guide to Common Crystallization Outcomes
| Outcome | Appearance | Probable Cause | Immediate Action |
|---|---|---|---|
| Clear Drop | No visible change; drop remains clear. | Protein remains in undersaturated or metastable zone; insufficient driving force for nucleation. | Increase protein concentration; screen with stronger precipitating agents. |
| Amorphous Precipitate | Granular or oily appearance; no birefringence. | Too high supersaturation, leading to rapid, disordered aggregation [47]. | Dilute precipitant concentration; use milder precipitating agents; employ additive screens. |
| Microcrystals | Tiny, birefringent particles (≤10 µm). | Nucleation is successful, but growth is hindered by impurities or unsuitable conditions. | Optimize seeding; reduce nucleation density; alter pH or temperature. |
| Phase Separation | Oily, spherical droplets forming in the drop. | Liquid-liquid phase separation often precedes crystal nucleation. | Allow more time; use as a positive indicator for nearby crystallization conditions. |
The following workflow provides a systematic approach for diagnosing and addressing these common crystallization failures, integrating the protocols detailed in subsequent sections.
Diagram 1: Crystallization Failure Decision Tree
Protocol 1: Reductive Alkylation for Salvaging Recalcitrant Proteins
For proteins that consistently fail to crystallize despite optimization, chemical modification of lysine residues via reductive alkylation is a powerful salvage technique. This method alters surface properties, reduces surface entropy, and can facilitate new crystal contacts [61].
Background and Principle
Reductive alkylation modifies solvent-exposed ε-amino groups of lysine residues, converting them from primary amines to secondary or tertiary amines. This reduces surface entropy and can modulate the protein's isoelectric point, thereby altering its crystallization behavior. The Midwest Center for Structural Genomics (MCSG) successfully used this approach to determine the structures of 12 unique proteins from a set of 180 that had previously failed, representing a significant increase in the structural determination success rate [61].
Step-by-Step Protocol
Materials:
- Purified protein (5–20 mg at 5–10 mg/ml)
- Dimethylamine-borane complex (reducing agent)
- Aldehyde source: Formaldehyde (methylation), Acetaldehyde (ethylation), or Acetone (isopropylation)
- Reaction buffer (e.g., 20 mM HEPES, pH 7.5)
Procedure:
- Preparation: Ensure the protein is in a buffer that does not contain primary amines (e.g., avoid Tris). A HEPES or phosphate buffer is suitable.
- Chilling: Cool all reagents and protein solution to 4°C.
- Reaction Setup: In sequence, add to the protein solution:
- The aldehyde source to a final concentration of 10-20 mM.
- The dimethylamine-borane complex to a final concentration of 20-40 mM.
- Incubation: Incubate the reaction mixture on ice or at 4°C for 2 hours.
- Quenching & Purification: Terminate the reaction by dialyzing the mixture into a suitable crystallization buffer or by using a desalting column.
- Concentration: Concentrate the alkylated protein and proceed with crystallization trials.
Validation: The efficiency of alkylation can be confirmed by mass spectrometry, which will show an increase in molecular mass (+14 Da per methyl group added).
Results and Efficacy
The table below summarizes the success rates of different alkylation strategies from the MCSG study, demonstrating that reductive methylation is the most efficient approach [61].
Table 2: Efficacy of Reductive Alkylation in Structural Determination [61]
| Alkylation Type | Proteins Treated | Macroscopic Crystals Harvested | Structures Solved |
|---|---|---|---|
| Methylation | 180 | 21 | 10 |
| Ethylation | 74 | 10 | 1 |
| Isopropylation | 21 | 4 | 1 |
| Total | 275 | 35 | 12 |
Protocol 2: Optimizing Sample Preparation to Prevent Precipitation
Amorphous precipitate often results from poor sample homogeneity or rapid, uncontrolled aggregation. Meticulous sample preparation is the first line of defense.
Assessing and Ensuring Sample Purity and Stability
- Purity: Aim for >95% homogeneity as assessed by SDS-PAGE and size-exclusion chromatography (SEC) [47]. Impurities act as nucleation points for disorder.
- Stability: Use differential scanning fluorimetry (DSF) to identify optimal buffer conditions, pH, and ligands that maximize thermal stability. Avoid phosphate buffers if possible, as they can form insoluble salts [47].
- Construct Design: Utilize predictive tools like AlphaFold to design constructs that remove flexible, disordered regions, reducing conformational heterogeneity that inhibits crystal lattice formation [47].
Evaluating and Achieving Monodispersity
- Dynamic Light Scattering (DLS): A monodisperse sample will show a single, sharp peak. Polydispersity indices >20-30% often correlate with poor crystallization outcomes.
- Size-Exclusion Chromatography Multi-Angle Light Scattering (SEC-MALS): This technique provides an absolute measure of molecular weight and homogeneity, confirming the protein is in the correct oligomeric state.
Managing Redox State
The choice of reducing agent is critical for proteins with cysteine residues. Consider the half-life of the reductant in your experimental conditions.
Table 3: Solution Half-Lives of Common Biochemical Reducing Agents [47]
| Chemical Reductant | Solution Half-life (hours) | Notes |
|---|---|---|
| Dithiothreitol (DTT) | 40 (at pH 6.5), 1.5 (at pH 8.5) | Short half-life at higher pH. |
| β-Mercaptoethanol (BME) | 100 (at pH 6.5), 4.0 (at pH 8.5) | Less efficient than DTT. |
| Tris(2-carboxyethyl)phosphine (TCEP) | >500 (pH 1.5–11.1) | Recommended for long-term stability; effective at a wide pH range. |
Protocol 3: Converting Microcrystals and Precipitate to Diffraction-Quality Crystals
When initial screens yield microcrystals or precipitate, the conditions are close to productive nucleation but require refinement.
Fine-Tuning Supersaturation
- For Precipitate: The supersaturation is too high. Systematically reduce the concentration of the precipitating agent (e.g., PEG or salt) in fine increments (e.g., 2-5% steps for PEG). Alternatively, perform a fine-screen around the precipitating condition by varying pH by ± 0.2-0.5 units.
- For Microcrystals: The supersaturation may be too high for controlled growth. Slightly decreasing the precipitant concentration can reduce the nucleation rate, allowing fewer nuclei to grow larger. Alternatively, slightly increasing the protein concentration can provide more nutrient for growth.
Seeding Strategies
Seeding is a highly effective technique for transferring crystal nuclei from a condition with high nucleation (microcrystals) to a new drop with optimal growth conditions (metastable zone).
- Harvest Seeds: Crush a microcrystal or a piece of a larger crystal from a similar condition in a stabilizing solution (e.g., mother liquor with a slightly lower precipitant concentration).
- Prepare Seed Stock: Serially dilute the crushed crystal slurry to achieve a range of seed densities.
- Transfer Seeds: Introduce a small volume of the seed stock into pre-equilibrated crystallization drops that are in the metastable zone (determined empirically as the highest precipitant concentration that does not lead to spontaneous nucleation). This provides growth sites without the energy barrier of nucleation.
Leveraging the "Genome Pool" Strategy
If one protein homologue consistently fails, a powerful high-throughput strategy is to screen multiple homologues simultaneously. Physicochemical properties that affect crystallizability can vary significantly even within a protein family, increasing the odds that at least one homologue will yield crystals [62].
The Scientist's Toolkit: Essential Reagents and Materials
Table 4: Key Research Reagent Solutions for Crystallization Salvage
| Reagent/Material | Function/Application | Example Use |
|---|---|---|
| Polyethylene Glycol (PEG) | Polymer for macromolecular crowding and salting-out; a ubiquitous precipitant. | Screens across a wide molecular weight (PEG 1K-20K) and concentration range (5-30%). |
| Ammonium Sulfate | Salt for salting-out proteins; promotes crystallization by dehydrating the protein solvation shell. | Commonly used in high concentrations for a wide variety of proteins. |
| 2-methyl-2,4-pentanediol (MPD) | Additive and precipitant; binds hydrophobic patches and affects hydration. | Used as a primary precipitant or additive to improve crystal morphology. |
| Reductive Alkylation Kit | Chemical modification of lysine residues to aid crystallization of recalcitrant proteins. | Salvage pathway for proteins that produce only precipitate or clear drops. |
| Tris(2-carboxyethyl)phosphine (TCEP) | Stable reducing agent to prevent disulfide bond formation and oxidation. | Preferred over DTT for long-term crystallization trials due to its long half-life. |
| Hampton Research Additive Screen | Library of small molecules to fine-tune crystal contacts and improve order. | Systematic screening of additives to optimize conditions yielding microcrystals. |
| Seed Bead (Hampton Research) | Commercial tool for easily harvesting and serial diluting of crystal seeds. | Standardizing and simplifying the microseeding protocol. |
Navigating common crystallization failures requires a systematic approach that moves from diagnosis to targeted intervention. By integrating rigorous sample preparation, employing strategic optimization of supersaturation, utilizing seeding techniques, and having robust salvage protocols like reductive alkylation in the toolkit, researchers can significantly increase the throughput and success of their crystal growth efforts. The strategies outlined here provide a concrete framework for transforming clear drops, precipitate, and microcrystals into diffraction-quality crystals, thereby accelerating structural discovery.
In the field of high-throughput protein crystallization screening, microfluidic technologies have emerged as transformative tools that enable researchers to overcome traditional limitations of sample volume and screening speed. Protein crystallization remains a critical bottleneck in structural biology and drug development, essential for determining three-dimensional protein structures via X-ray crystallography [6]. Conventional methods, such as vapor diffusion and batch crystallization in microtiter plates, typically consume microliter volumes of precious protein samples and can screen only a limited number of conditions [63]. The integration of microfluidics has revolutionized this landscape by facilitating the manipulation of nanoliter to picoliter fluid volumes, allowing for unprecedented miniaturization and parallelization of crystallization trials [63] [64]. This advancement is particularly valuable in structural genomics initiatives and pharmaceutical development, where the demand for rapid protein structure determination continues to accelerate alongside discoveries from genomic and metagenomic surveillance programs [50].
Quantitative Advantages of Microfluidic Platforms
The transition from conventional crystallization methods to microfluidic approaches yields significant quantitative benefits, particularly in sample consumption and experimental throughput. The data in Table 1 illustrates these advantages across different crystallization platforms.
Table 1: Comparison of Crystallization Platform Capabilities
| Platform Type | Typical Volume Range | Throughput (Conditions per Run) | Key Advantages |
|---|---|---|---|
| Batch Crystallizers | 0.1 - 1 L | Limited | Suitable for kinetic studies, scalable |
| Mini-Batch Crystallizers | ~1 mL | ~16 conditions simultaneously | Enables polymorphism and morphology studies |
| Microtiter Plates | 1 μL - 1 mL | Up to 192 conditions (≈10,000/day) | Fully automated screening |
| Microfluidic Devices | < 1 μL | Up to 2,500 conditions | High-throughput, stationary environment for kinetic studies |
| Nanofluidic Devices | < 100 nL | Developing technology | Novel phase measurement capabilities |
Data compiled from [63]
As evidenced in Table 1, microfluidic devices operate with volumes below 1 μL, dramatically reducing protein sample requirements while enabling the screening of thousands of crystallization conditions in a single experiment [63]. This miniaturization is particularly crucial for challenging targets such as membrane proteins or proteins available only in limited quantities [6]. The exceptional surface-to-volume ratios in microfluidic systems enable superior control over mass and heat transfer, leading to more precise manipulation of crystallization kinetics and outcomes [63].
Key Microfluidic Modalities and Applications
Droplet-Based Microfluidics
Droplet-based microfluidic platforms generate discrete nanoliter volumes that function as independent micro-reactors, greatly enhancing screening efficiency. One innovative approach combines microfluidic mixing with droplet generation to create populations of identical droplets for crystallization trials [64]. Researchers have successfully utilized this technology to rigorously quantify nucleation parameters, demonstrating that functionalized nanoparticles can reduce induction time by up to 7-fold and increase nucleation rates by 3-fold compared to control environments [64]. This platform enables time-course imaging of individual droplets to determine induction times and nucleation rates, providing valuable kinetic data previously difficult to obtain with conventional methods.
Digital Microfluidics (DMF)
Digital microfluidics (DMF) represents another powerful approach wherein discrete droplets are electrically manipulated across an array of electrodes. Recent advances have addressed the challenge of processing large sample volumes (up to 100 μL) with low bead counts (as few as 5,000) for ultrasensitive single molecule array (Simoa) digital protein assays [65]. The development of "virtual channels" and "densifying electrodes" enables efficient bead recovery and solution exchange, facilitating the miniaturization of protein detection assays that traditionally require large sample volumes [65]. This capability is particularly valuable for detecting low-abundance protein biomarkers in clinical and biomedical applications.
Microbatch Under Oil
The microbatch-under-oil method has been widely adopted in high-throughput crystallization centers, with the National Crystallization Center reporting the setup of over 25 million crystallization experiments on more than 18,000 biological macromolecules [66]. This approach involves dispensing nanoliter-scale protein-precipitant mixtures under an oil layer to prevent evaporation, allowing efficient sampling of chemical parameter space across 1,536 non-redundant crystallization conditions [66]. When combined with state-of-the-art imaging techniques including brightfield microscopy and Second-Order Non-linear Imaging of Chiral Crystals (SONICC), this method enables the detection of crystals that might be missed by visual inspection alone [66].
Experimental Protocols
Protocol 1: High-Throughput Screening via Droplet Microfluidics
Objective: To identify initial protein crystallization conditions using a droplet-based microfluidic platform.
Materials:
- Purified protein sample (>5 mg/mL)
- Crystallization screening solutions
- Droplet generation oil and surfactants
- Microfluidic device (droplet generator and mixer)
- Microscopy system with camera and temperature control
Procedure:
- Device Preparation: Prime the microfluidic channels with carrier oil and surfactant solution to ensure stable droplet formation.
- Solution Loading: Load the protein solution and crystallization reagent solutions into separate syringes connected to the microfluidic device.
- Droplet Generation: Set flow rates to achieve desired droplet size (typically 10-100 nL). The platform combines a microfluidic mixer and emulsion generator to produce a population of identical, independent droplets containing protein and precipitant at the target concentration [64].
- Incubation: Collect droplets in a capillary tube or well plate and maintain at constant temperature for crystallization.
- Imaging and Analysis: Image droplets at regular intervals using a microscope connected to a camera with polarizers. Analyze the fraction of clear drops at each time point to determine induction time and nucleation rate [64].
Protocol 2: Optimization via Vapor Diffusion in Microfluidic Devices
Objective: To optimize crystallization conditions identified from initial screens.
Materials:
- Protein solution
- Optimized crystallization solutions based on initial hits
- Microfluidic vapor diffusion chips
- Sealing tape or oil for evaporation control
Procedure:
- Chip Loading: Pipette nanoliter volumes of protein solution and crystallization solutions into adjacent wells on the microfluidic chip.
- Sealing: Seal the device to control vapor diffusion between wells.
- Equilibration: Allow the device to equilibrate at constant temperature, enabling water transfer from the protein solution to the reservoir solution to gradually increase protein concentration [64].
- Monitoring: Observe crystal formation over time using built-in imaging capabilities or off-line microscopy.
- Harvesting: For X-ray crystallography, retrieve crystals directly from the device using specialized tools or by breaking the device.
Protocol 3: Integrating Functionalized Nanoparticles to Enhance Crystallization
Objective: To utilize bioconjugate-functionalized nanoparticles to improve crystallization success.
Materials:
- Protein sample (e.g., lysozyme or insulin)
- Functionalized nanoparticles (e.g., 16 μL added to 1 mL of protein solution) [64]
- Sodium chloride in sodium acetate buffer
- Microfluidic crystallization device
Procedure:
- Solution Preparation: Prepare protein solution at target concentration (e.g., 10-50 mg/mL) in appropriate buffer.
- Nanoparticle Addition: Add functionalized nanoparticles to the protein solution to achieve target concentrations and supersaturation levels [64].
- Droplet Generation: Utilize the microfluidic platform to generate emulsion droplets containing the protein-nanoparticle mixture.
- Incubation and Monitoring: Maintain droplets at constant temperature and image at regular intervals.
- Data Analysis: Compare induction times and nucleation rates between nanoparticle-enhanced and control experiments to quantify enhancement effects.
Workflow and Signaling Pathways
The following diagrams illustrate the experimental workflow for high-throughput microfluidic crystallization screening and the decision process for method selection based on protein characteristics.
Diagram 1: High-throughput microfluidic crystallization screening workflow.
Diagram 2: Decision process for microfluidic crystallization method selection.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Research Reagents and Materials for Microfluidic Crystallization
| Reagent/Material | Function | Example Application |
|---|---|---|
| Tetronic 90R4 Surfactant | Stabilizes droplets in microfluidic channels; prevents fusion | Used in digital microfluidics at 0.01-0.1% (w/v) concentration [65] |
| Superparamagnetic Beads (2.7 μm) | Solid support for protein capture and manipulation | Enables single molecule array (Simoa) digital protein assays [65] |
| Bioconjugate-functionalized Nanoparticles | Enhances nucleation rates; reduces induction time | Up to 7-fold decrease in induction time and 3-fold increase in nucleation rate [64] |
| pMCSG53 Vector | Protein expression vector with cleavable N-terminal hexa-histidine tag | High-throughput transformation, expression, and solubility screening [50] |
| Crystallization Reagents & Screens | Provides diverse chemical space for crystallization trials | 1,536 non-redundant conditions in high-throughput screening [66] |
| Microfluidic Chips | Platform for fluid manipulation at microscale | Droplet generation, mixing, and incubation [63] [64] |
Microfluidic technologies have fundamentally transformed the landscape of high-throughput protein crystallization by dramatically reducing sample volume requirements while exponentially increasing screening capabilities. The ability to conduct thousands of experiments with nanoliter-scale consumption of precious protein samples has accelerated structural genomics initiatives and drug discovery pipelines. As these technologies continue to evolve, integrating with artificial intelligence for experimental design and analysis [9], and expanding into new applications such as membrane protein crystallization [6], their impact on structural biology will undoubtedly grow. The protocols and methodologies outlined in this application note provide researchers with practical frameworks to leverage the power of microfluidics in their protein crystallization endeavors, ultimately contributing to the expanded understanding of protein structure and function that underpins modern drug development.
Validating Success and Navigating the Evolving Technological Landscape
Structural genomics pipelines have revolutionized the process of protein structure determination by integrating high-throughput automation, advanced computational prediction, and systematic experimental screening. The primary objective of these integrated pipelines is to overcome the historically low success rates in protein crystallization, which typically range between 2% and 10% [67] [68]. This high attrition rate, coupled with the extensive costs associated with failed experiments, has driven the development of sophisticated target selection and prioritization strategies. By benchmarking these methodologies, researchers can identify the most effective approaches for predicting crystallization propensity, ultimately accelerating structural biology and drug discovery efforts. The continuous evolution of these pipelines is evidenced by the projected growth of the protein crystallization market to $2.8 billion by 2029, reflecting the increasing importance of efficient structural biology techniques in biomedical research [9].
Computational Prediction: Benchmarking Protein Language Models
The integration of artificial intelligence, particularly protein language models (PLMs), represents a paradigm shift in predicting protein crystallization propensity. These models leverage self-supervised learning on millions of protein sequences to capture intricate patterns and physicochemical properties correlated with crystallizability.
Performance Benchmarking of PLMs
Recent benchmarking studies demonstrate that classifiers built on embeddings from specific PLMs significantly outperform traditional sequence-based methods. The table below summarizes the performance gains achieved by top-performing models on independent test sets.
Table 1: Performance comparison of protein language models for crystallization propensity prediction
| Model | Key Architecture | Performance Gain over SOTA | Key Evaluation Metrics |
|---|---|---|---|
| ESM2 (30 & 36 layers) | Transformer-based | 3-5% gain over DeepCrystal, ATTCrys, CLPred [67] | AUPR, AUC, F1 Score |
| LightGBM Classifier | Gradient Boosting on ESM2 embeddings | Most effective classifier [67] | AUPR, AUC, F1 Score |
| DeepCrystal | Convolutional Neural Network (CNN) | Baseline method [67] | - |
| ATTCrys | Multi-scale, multi-head self-attention CNN | Baseline method [67] | - |
| CLPred | Bidirectional LSTM (BLSTM) | Baseline method [67] | - |
The benchmarking revealed that LightGBM or XGBoost classifiers utilizing average embedding representations from ESM2 models with 30 and 36 transformer layers (150 million to 3 billion parameters) achieved superior performance [67]. These models excel at capturing long-range dependencies and subtle sequence patterns that traditional feature-engineering methods miss.
Experimental Protocol: Implementing PLM-Based Prediction
For researchers seeking to implement these computational predictors, the following protocol outlines the standard workflow:
- Sequence Input and Tokenization: Provide the target protein's amino acid sequence as input. The model's tokenizer encodes the sequence into an ingestible numerical representation, ( t(x) \in \mathbb{R}^L ), where ( L ) is the sequence length [67].
- Embedding Generation: Process the encoded sequence through the pre-trained PLM (e.g., ESM2, Ankh, ProtT5-XL). The final transformer layer generates a high-dimensional embedding representation for the entire protein, which encapsulates meaningful inter-residue relationships [67].
- Classifier Prediction: Input the generated embedding vector into a trained LightGBM or XGBoost classifier. The classifier outputs a numerical propensity score indicating the likelihood of successful crystallization [67].
- Validation and Interpretation: Compare the prediction against established benchmarks. The TRILL platform democratizes access to these models, eliminating the requirement for advanced computational skills [67].
Experimental Screening: Methodologies and Reagents
While computational prediction efficiently prioritizes targets, experimental screening remains indispensable for discovering specific crystallization conditions. Several optimized screen formulations have been developed to maximize the probability of success.
Benchmarking Screening Methodologies
The efficiency of a crystallization screen is measured by its ability to identify initial crystallization "hits" with the fewest experiments. The table below compares several widely used 96-condition screens.
Table 2: Comparison of high-throughput protein crystallization screens
| Screen Name | Formulation Strategy | Key Components | Application Notes |
|---|---|---|---|
| LMB Sparse Matrix | Empirical selection of successful conditions [69] | PEGs (46%), Salts, Volatiles, pH 5.0-7.9 [69] | Optimized for soluble proteins and complexes [69] |
| Pi Incomplete Factorial | Maximally diverse combinations via Pi sampling [69] | User-defined reagents (e.g., Pi-PEG for GPCRs) [69] | Customizable based on protein properties [69] |
| MORPHEUS | Grid screen integrating mixes of additives [69] | Precipitant mixes, buffer systems, additive mixes [69] | Incorporates cryo-protection and heavy atoms for phasing [69] |
| ANGSTROM | Optimization screen | Polyols | Used for crystal optimization [69] |
Efficiency analyses indicate that screening protocols utilizing subsets of related conditions can identify initial hits more efficiently than purely random sampling, making them ideal for scenarios with limited protein sample [70].
Research Reagent Solutions
Successful crystallization experiments rely on a core set of reagents and instruments. The following table details essential components.
Table 3: Essential research reagents and equipment for high-throughput crystallization
| Item Category | Specific Examples | Function in Crystallization |
|---|---|---|
| Precipitants | Polyethylene Glycols (PEGs), Ammonium Sulfate, MPD [69] [71] | Drives protein supersaturation by excluding volume [69] |
| Buffers | HEPES, Tris, MES, Acetate [71] | Controls pH of the crystallization solution [71] |
| Additives | Salts, Metals, Ligands, Detergents [69] | Fine-tunes protein interactions and stability [69] |
| Consumables | 24-well VDX Plates, 96-well Intelli-Plates, Siliconized Cover Slips [71] | Hardware for vapor diffusion experiments [71] |
| Automation | Crystal Gryphon Automated Liquid Handler [71] | Enables high-throughput, nanoliter-volume setup [71] |
Integrated Structural Genomics Pipeline
The most successful structural genomics centers do not rely on a single method but integrate computational prediction with robust experimental screening in a synergistic pipeline. The workflow below visualizes this integrated approach.
Diagram 1: Integrated structural genomics workflow.
Protocol: High-Throughput Crystallization Screening
The following detailed protocol is adapted for an automated liquid handling system, such as the Crystal Gryphon.
Protein Sample Preparation:
- Purification: Ensure protein is >90% pure, as evaluated by SDS-PAGE or mass spectrometry [71].
- Concentration: Concentrate protein to a typical stock concentration of 10-20 mg/mL using an appropriate concentrator [71].
- Clarification: Remove dust and precipitate by centrifugation at 14,000 x g for 5-10 minutes at 4°C [71].
Automated Tray Setup (e.g., Crystal Gryphon):
- System Preparation: Power up the instrument and ensure wash stations have adequate water. Load the software and connect to the instrument [71].
- Reagent Loading:
- Position a deep-well block containing the screening solutions (e.g., a commercial 96-condition screen) on the stage. Remove the sealing tape [71].
- Place an empty crystallization plate (e.g., a 96-well Art Robbins Intelli-Plate) on the stage [71].
- Load a PCR tube containing at least 80-100 µL of clarified protein solution. Open the lid before placing it on the stage [71].
- Protocol Execution:
- Select the appropriate dispensing protocol (e.g., "2-drop Screen" for two protein concentrations).
- Initiate the run. The instrument will automatically dispense reservoir solution, then combine nanoliter volumes of screen solution and protein in the crystallization wells [71].
- Sealing and Storage: Once dispensing is complete, the plate is automatically sealed. Manually transfer plates to a stable temperature environment (e.g., 4°C or 20°C) and leave undisturbed [71].
Imaging and Hit Detection:
- Use an automated imaging system to regularly monitor drops for crystal growth. Initial examination should occur within 24 hours [71].
Benchmarking studies across structural genomics pipelines consistently reveal that an integrated strategy is paramount for maximizing success rates. Computational pre-screening using top-performing Protein Language Models like ESM2 can enhance experimental throughput by 3-5% by prioritizing the most promising targets [67]. Subsequently, employing efficient, methodical experimental screens such as sparse matrix or incomplete factorial screens systematically navigates the complex crystallization landscape. As the field advances with automation and AI, the continued benchmarking and refinement of each component within this integrated pipeline will remain fundamental to solving challenging protein structures, thereby empowering drug discovery and deepening our understanding of biological mechanisms.
Structural biology has been transformed by technological revolutions in experimental and computational methods. The integration of high-throughput X-ray crystallography, cryo-electron microscopy (cryo-EM), and artificial intelligence (AI)-based modeling has created a powerful multidisciplinary framework for elucidating protein structures at unprecedented speed and resolution. Within drug discovery pipelines, these technologies provide complementary insights into molecular interactions, enabling targeted therapeutic development [72]. This document provides application notes and experimental protocols for these technology platforms, contextualized within high-throughput protein crystallization research.
The convergence of these fields is particularly evident in natural product-based drug discovery, where high-throughput crystallography directly captures bioactive compounds from unpurified samples [73]. Simultaneously, automated machine learning approaches like ModelAngelo now build atomic models from cryo-EM maps with accuracy rivaling human experts [74]. These advances occur alongside a growing protein crystallization market, projected to reach $2.8 billion by 2029, driven by automation, AI integration, and next-generation X-ray technologies [9].
Technology Platform Comparisons
Performance Metrics and Applications
Table 1: Comparative analysis of structural biology platforms for high-throughput applications
| Parameter | X-ray Crystallography | Cryo-EM | AI Modeling |
|---|---|---|---|
| Typical Resolution Range | Atomic (1-2 Å) | Near-atomic to atomic (1.5-4 Å) | Accuracy comparable to experimental methods for well-folded domains [72] [74] |
| Sample Requirements | High purity, crystallization | Low sample quantity, tolerance for heterogeneity | Amino acid sequence only |
| Throughput Capacity | High with automation | Moderate to high | Very high (proteome-scale) |
| Membrane Protein Suitability | Challenging, requires LCP methods | Excellent | Good for single chains, limited for complexes |
| Dynamic Information | Time-resolved variants possible | Limited | Limited to static predictions |
| Equipment Cost | High ($500K-$2M+) | Very High ($1M-$5M+) | Low (computational infrastructure) |
| Automation Level | Extensive automation available | Growing automation | Fully automated |
| Key Strengths | Gold standard resolution, high throughput | Native sample visualization, membrane proteins | Speed, scalability, no sample needed |
Economic and Throughput Considerations
Table 2: Economic and operational characteristics for high-throughput structural biology
| Characteristic | X-ray Crystallography | Cryo-EM | AI Modeling |
|---|---|---|---|
| Market Size (2024) | $1.62B (crystallization overall) [9] | N/A | N/A |
| Projected Growth | 11.5% CAGR to $2.8B by 2029 [9] | Rapid adoption | Exponential growth |
| Primary End-Users | Pharmaceutical & biotechnology companies, academic institutes [9] | Academic, pharmaceutical | All sectors |
| Automation Level | Extensive (liquid handlers, imagers) [8] | Growing (grid preparation, screening) | Fully automated |
| Data Analysis | AI-assisted scoring [8] | ModelAngelo for automated building [74] | Integrated pipelines |
| Key Limitations | Crystallization bottleneck, membrane proteins | Resolution variability, cost | Limited conformational diversity |
Integrated Experimental Protocols
High-Throughput Protein Crystallization Screening
Objective: Rapid identification of crystallization conditions for protein-ligand complexes using automated workflows.
Materials and Equipment:
- Purified target protein (>95% purity)
- Crystallization screens (commercial or custom)
- OPENTRONS-2 liquid handling robot or Tecan Freedom EVO 200 [75] [27]
- Formulatrix NT8 Drop Setter [8]
- Formulatrix Rock Imager with UV, MFI, or SONICC capabilities [8]
- 24-well sitting drop plates or 96-well format plates
- Humidity-controlled incubation environment
Procedure:
- Protein Preparation:
- Concentrate protein to 5-20 mg/mL in appropriate buffer
- Centrifuge at 14,000 × g for 10 minutes to remove aggregates
- Maintain at 4°C throughout process
Automated Screen Preparation:
- Program liquid handler to dispense crystallization reagents
- Use Formulator Screen Builder for custom condition generation [8]
- Transfer 50-100 μL of each condition to reservoir wells
Drop Setup:
Incubation and Monitoring:
- Seal plates with clear adhesive
- Incubate at constant temperatures (4°C, 20°C, or 37°C)
- Image plates automatically using Rock Imager at 24-hour intervals
- Apply multiple imaging modalities (brightfield, UV, SONICC) [8]
Hit Identification and Optimization:
- Use AI-based autoscoring (Sherlock model) to identify crystals [8]
- Classify hits based on crystal morphology and size
- Optimize hits via additive screening or seeding
Troubleshooting Notes:
- For low yields, consider microbatch under oil or free-interface diffusion
- If crystals don't diffract well, optimize cryoprotection or try micro-seeding
- For membrane proteins, employ fluorescence recovery after photobleaching (FRAP) to screen LCP conditions [8]
Integrated Cryo-EM and AI Modeling Pipeline
Objective: Determine atomic structures of protein complexes using cryo-EM with AI-assisted model building.
Materials and Equipment:
- Vitrobot or equivalent plunge freezer
- Transmission electron microscope with direct electron detector
- ModelAngelo software (version 1.0 or higher) [74]
- Protein samples at 0.5-3 mg/mL in minimal buffer
- UltrauFoil or Quantifoil grids
Procedure:
- Sample Preparation and Grid Freezing:
- Apply 3-5 μL protein to freshly plasma-cleaned grids
- Blot for 2-6 seconds at 100% humidity
- Plunge freeze in liquid ethane cooled by liquid nitrogen
- Screen grids for ice thickness and particle distribution
Data Collection:
- Collect 2,000-5,000 micrographs at nominal 130,000× magnification
- Use dose-fractionation mode with total dose of 40-60 e⁻/Ų
- Implement energy filtering if available
- Collect movies with 40-50 frames per exposure
Image Processing and Map Generation:
- Perform beam-induced motion correction and dose weighting
- Estimate contrast transfer function parameters
- Execute particle picking, extraction, and 2D classification
- Generate initial model ab initio or from AlphaFold prediction [72]
- Refine 3D reconstruction with imposed symmetry if applicable
AI-Assisted Model Building:
- Input final cryo-EM map and protein sequences into ModelAngelo [74]
- Run Cα and phosphate position prediction using convolutional neural network
- Apply graph neural network to optimize residue positions and identities
- Incorporate sequence information via ESM-1b protein language model [74]
- Generate atomic model with side-chain placements
Model Validation and Refinement:
- Compute model-to-map correlation metrics
- Validate geometry using MolProbity or similar tools
- Perform iterative manual adjustment in Coot if needed
- Finalize models with phenix.realspacerefine or equivalent
Cross-Platform Integration for Drug Discovery
Objective: Combine crystallography, cryo-EM, and AI to accelerate structure-based drug design.
Procedure:
- Initial Target Assessment:
- Use AlphaFold predictions to assess crystallizability [72]
- Identify flexible regions requiring cryo-EM approach
- Generate structural models for virtual screening
Multi-Technology Structure Determination:
- Pursue parallel crystallization and cryo-EM screening
- Employ high-throughput crystallography for ligand complexes [73]
- Use cryo-EM for large complexes and membrane proteins
Data Integration and Modeling:
- Combine AlphaFold predictions with experimental maps [72]
- Build composite models using flexible fitting
- Validate against experimental data from both techniques
Drug Discovery Applications:
- Perform fragment screening using crystallographic capture [73]
- Characterize drug-bound complexes with cryo-EM
- Utilize AI-predicted structures for targets resistant to experimental methods
Research Reagent Solutions and Materials
Table 3: Essential research reagents and materials for high-throughput structural biology
| Item | Function | Example Products/Suppliers |
|---|---|---|
| Liquid Handling Robots | Automated dispensing of crystallization trials | Tecan Freedom EVO 200, Opentrons-2, Formulatrix NT8 [75] [8] [27] |
| Crystallization Screens | Pre-formulated condition matrices | NeXtal crystallization suites (Qiagen), Molecular Dimensions screens [9] [75] |
| Automated Imagers | High-throughput crystal detection and monitoring | Formulatrix Rock Imager series (UV, MFI, SONICC capabilities) [8] |
| Direct Electron Detectors | High-resolution cryo-EM data collection | Falcon, K2, and K3 detectors [72] |
| AI Modeling Software | Automated structure prediction and model building | ModelAngelo, AlphaFold 2/3, RoseTTAFold [72] [74] |
| Laboratory Information Systems | Managing crystallization experimental data | Rock Maker crystallization software [8] |
| Cryo-EM Grids | Sample support for cryo-EM | UltrauFoil, Quantifoil, C-flat |
The synergistic integration of X-ray crystallography, cryo-EM, and AI modeling platforms has created unprecedented opportunities for structural biology and drug discovery. As evidenced by the protocols outlined herein, automated workflows now enable rapid transition from protein targets to atomic structures, significantly accelerating research timelines. The continued development of integrated approaches, particularly between cryo-EM and X-ray sciences, promises enhanced capabilities for capturing molecular dynamics and facilitating targeted drug design [76].
For researchers engaged in high-throughput protein crystallization screens, the strategic implementation of complementary technologies is essential. AI-guided experimental design, automated crystal detection, and machine learning-assisted model building represent the current state-of-the-art, with further advances anticipated as these fields continue to converge. The result is an increasingly powerful toolkit for elucidating protein structure-function relationships and advancing therapeutic development.
Protein crystallization is a critical step in structural biology, enabling researchers to determine the three-dimensional structures of proteins via techniques like X-ray crystallography [14]. For researchers and drug development professionals, optimizing the economic and operational aspects of high-throughput crystallization screens is paramount to accelerating discovery while managing resources. The global protein crystallization market, valued at between $6.80 billion and $7.72 billion in 2025, is growing at a significant compound annual growth rate (CAGR) of 11.3% to 14.00%, underscoring the technique's expanding role in biotechnology and pharmaceutical development [16] [10]. This application note provides a detailed economic and operational analysis of high-throughput protein crystallization screening, featuring structured cost data, actionable protocols, and strategic guidance to inform laboratory planning and investment.
Market Context and Key Drivers
The growth of the protein crystallization market is propelled by several key factors. The rising demand for biopharmaceuticals, such as monoclonal antibodies and engineered enzymes, requires atomic-level structural data for regulatory filings and drug development pipelines [14] [10]. Furthermore, technological advancements in automation, microfluidics, and artificial intelligence (AI) are transforming traditional crystallization workflows from empirical, trial-and-error processes into efficient, data-driven operations [14] [16]. These innovations are crucial for addressing persistent operational challenges, including the high capital costs of equipment and a shortage of highly skilled crystallographers [14]. The table below summarizes the quantitative market landscape.
Table 1: Global Protein Crystallization Market Outlook
| Metric | Value (2024/2025) | Projected Value (2032) | CAGR | Primary Drivers |
|---|---|---|---|---|
| Market Size | $7.72 billion (2025) [16] | $19.41 billion [16] | 14.00% [16] | Rising biopharma R&D, adoption of protein therapeutics [14] [10] |
| Alternative Market Size | $1.81 billion (2025) [10] | $2.78 billion (2029) [10] | 11.3% [10] | Demand for targeted treatments, high-throughput screening [10] |
| Fastest-growing Product Segment | Software & Services [14] | - | 12.19% [14] | Need for AI-based structure prediction, data analysis [14] [16] |
| Fastest-growing End User | Contract Research Organizations (CROs) [14] | - | 10.24% [14] | Outsourcing by small- and mid-sized biotechs [14] |
| Fastest-growing Region | Asia-Pacific [14] | - | 10.05% [14] | Government investment in synchrotrons, life science infrastructure [14] |
Economic Analysis: Cost Considerations
A thorough economic analysis must account for both significant capital expenditures and ongoing operational costs. The following table breaks down the key economic factors impacting high-throughput crystallization workflows.
Table 2: Economic and Operational Factor Analysis
| Factor | Economic/Operational Impact | Strategic Mitigation |
|---|---|---|
| Capital Equipment | High-cost instruments: X-ray diffractometers/cryo-EM rigs can exceed $7 million per unit [14]. | Utilize academic service centers (~$150–450/sample), leasing equipment, partnering with CROs [14] [16]. |
| Skilled Expertise | Shortage of trained crystallographers creates bottlenecks; demand outstrips supply [14]. | Invest in training, utilize AI software to augment human expertise, outsource to specialized CROs [14] [16]. |
| Throughput & Efficiency | Traditional screening requires hundreds/thousands of conditions, consuming time/reagents [77] [78]. | Adopt microfluidic platforms (reduce sample volume, screen 1000s of conditions in 30min) and AI-guided screening [14]. |
| Supply Chain & Tariffs | 2025 U.S. tariff adjustments increase costs of reagents/instruments,延长lead times [16]. | Diversify suppliers, establish long-term agreements, explore regional manufacturing, bulk purchasing consortia [16]. |
Operational Analysis: Throughput and Workflow Solutions
Operational efficiency in high-throughput crystallization is achieved through technological integration and workflow optimization.
The Evolution of Workflow Throughput
Modern workflows have evolved from manual, low-throughput methods to highly automated and miniaturized processes. Liquid handling robots and crystallization imaging systems now enable the setup and monitoring of thousands of trials in parallel [14] [16]. This automation is complemented by microfluidic platforms, which reduce sample volume requirements by an order of magnitude and allow for the screening of thousands of conditions within approximately 30 minutes [14]. The integration of AI-powered software for crystal detection and condition prediction further transforms the operational paradigm from one of trial-and-error to a data-driven, iterative process that accelerates hit identification and optimization [14] [16].
Associative Experimental Design (AED) Protocol
To maximize the efficiency of screening campaigns, the Associative Experimental Design (AED) method provides an intelligent, results-driven approach to optimizing crystallization conditions [77] [78].
1. Initial Screening:
- Perform a broad, sparse-matrix initial screen (e.g., 96-196 conditions) against your purified protein target.
- Incubate the trials under controlled temperature conditions and monitor results regularly using an automated imaging system.
2. Condition Scoring:
- Score the outcomes of the initial screen using a standardized system like the Hampton Score, where scores of 4 to 7 typically indicate crystalline outcomes or promising hits [78].
- Record the specific chemical condition (precipitant, salt, buffer, additive) for each well.
3. AED Data Analysis:
- The AED algorithm analyzes the initial screening results to identify the specific chemical factors (e.g., type and concentration of precipitant, specific salts, pH) that are statistically associated with high-scoring outcomes.
- The method effectively prioritizes reagents and can eliminate combinations known to be prohibitive [77].
4. Generation of Optimized Conditions:
- Based on the analysis, AED generates a list of novel, focused candidate crystallization cocktails that emphasize the factors linked to success.
- This list is typically smaller and more targeted than the initial broad screen.
5. Experimental Validation:
- Set up a second-round of crystallization trials using the novel conditions proposed by the AED analysis.
- Validation on proteins like Nucleoside diphosphate kinase has yielded a significant increase in high-scoring crystalline conditions (e.g., 20 conditions) compared to initial screens [77] [78].
The following workflow diagram illustrates the high-throughput screening process, highlighting the critical feedback loop for optimization.
The Scientist's Toolkit: Essential Research Reagent Solutions
A successful high-throughput crystallization screen relies on a foundation of key reagents and instruments. The following table details essential materials and their functions within the workflow.
Table 3: Key Research Reagent Solutions for Crystallization Screening
| Item | Function | Application Note |
|---|---|---|
| Crystallization Reagents & Screens | Sparse-matrix or grid screens containing precipitants, salts, buffers, and additives to sample a wide chemical space. | Formulations like sodium-malonate that act as both precipitant and cryoprotectant can streamline workflows [14]. |
| Microplates & Crystallization Plates | Miniaturized platforms (e.g., 96-well, 384-well) for setting up nanoliter-scale vapor-diffusion trials. | Compatible with robotic liquid handlers (e.g., SPT Labtech's mosquito crystal) to maximize throughput and minimize reagent use [14] [10]. |
| Liquid Handling Systems | Automated robots and microfluidic platforms for precise, nanoliter-scale dispensing of protein and reagent solutions. | Microfluidic chips can reduce sample needs by an order of magnitude and dramatically speed up screening [14]. |
| Crystallization Imaging Systems | Automated microscopes for high-throughput, time-lapsed monitoring of crystal growth in incubation chambers. | Integrated AI algorithms enable automated crystal detection, reducing manual oversight and error rates [16]. |
| Cryoprotectants | Chemicals (e.g., glycerol, ethylene glycol) used to protect crystals from ice damage during cryo-cooling prior to X-ray data collection. | An essential final step before harvesting crystals for synchrotron data collection [14] [10]. |
Strategic Implementation Guidance
For research leaders, strategic implementation is key to leveraging these analyses for operational success. Prioritize Integrated Automation by investing in modular robotics platforms that combine liquid handling, incubation, and imaging with AI-driven condition optimization to reduce trial iterations and accelerate time-to-structure [16]. Embrace a Hybrid Sourcing Model to mitigate capital expenditure and expertise shortages; this involves maintaining core internal capability for IP-sensitive projects while strategically outsourcing to specialized CROs for specific targets or to manage workflow overflow [14] [16]. Commit to Data-Driven Optimization by implementing software platforms that utilize machine learning and methods like Associative Experimental Design, which analyzes preliminary results to intelligently guide subsequent screening rounds, maximizing the output from every experiment [77] [16] [78].
The field of high-throughput protein crystallization is undergoing a transformative shift, driven by the convergent evolution of artificial intelligence (AI) and microfluidic miniaturization. These technologies are collectively addressing the most persistent challenges in structural biology: the inefficiency, high resource consumption, and low throughput of traditional crystallization methods. For researchers and drug development professionals, this synergy is not merely an incremental improvement but a fundamental redesign of the crystallization workflow. It enables a shift from labor-intensive, empirical screening to a predictive, data-driven paradigm that accelerates the path from protein purification to high-resolution structural data.
The commercial and research landscapes reflect this rapid adoption. The protein crystallization market, valued at $1.62 billion in 2024, is projected to grow to $2.8 billion by 2029, representing a robust compound annual growth rate (CAGR) of 11.5% [9]. This growth is fueled by the rising demand for protein-based therapeutics and the integration of cutting-edge technologies that enhance efficiency and success rates [79] [9]. This application note details the specific protocols and tools that are leveraging this technological convergence to redefine high-throughput screening.
Table 1: Key Market and Technology Drivers in Protein Crystallization
| Aspect | Current Impact | Future Outlook |
|---|---|---|
| Market Size (2024) | USD 1.62 Billion [9] | USD 2.8 Billion by 2029 [9] |
| Primary Growth Driver | Demand for biologics and targeted therapeutics [9] | Expansion into structural genomics and personalized medicine [79] |
| AI Integration | Machine learning for crystal scoring and condition prediction [80] | AI-driven predictive modeling for crystallization condition optimization [81] |
| Microfluidic Trends | Miniaturization to reduce sample consumption [82] | Integrated systems for in situ analysis and serial crystallography [82] [83] |
The AI Revolution in Crystallization Workflows
Artificial intelligence is revolutionizing protein crystallization by bringing unprecedented levels of automation and predictive power to experimental processes. The core of this revolution lies in the deployment of sophisticated machine learning models that automate scoring and enhance experimental design.
Automated Crystal Detection with MARCO
A critical bottleneck in high-throughput crystallization is the manual scoring of thousands of crystallization drops, a process that is both time-consuming and subjective. The Machine Recognition of Crystallization Outcomes (MARCO) system, integrated into the Rock Maker software platform, addresses this directly [80]. This machine learning algorithm automatically classifies drop images as Crystal, Precipitate, Clear, or Other.
- Performance Metrics: MARCO can score a 96-well, single-drop plate in under 3 minutes with an reported accuracy of approximately 94% [80]. This drastically reduces the analysis time from days to hours for large screening campaigns.
- Functionality: The system provides a probability score for each potential outcome, allowing researchers to prioritize hits based on confidence levels. This is a key feature of Rock Maker's dynamic image viewing and scoring module [80].
AI-Enhanced Optimization and Future Trends
Beyond scoring, AI is being leveraged to optimize experiments. Rock Maker's Iterative Screen Optimization (ISO) is a highly automated process that uses initial user scores to automatically generate follow-up experiments. It systematically adjusts precipitant concentrations to navigate the protein solubility curve and pinpoint the nucleation zone [80]. The future of AI in this field, as previewed in upcoming workshops, involves developing models that can predict crystallization conditions before a single experiment is set up, and brainstorming the next generation of AI tools to answer complex experimental questions [81].
Microfluidic Miniaturization for Sample Conservation
Parallel to the AI revolution, microfluidic technologies are addressing the critical issue of sample consumption. Precious, hard-to-express proteins are no longer a prohibitive factor for structural studies, thanks to devices that minimize volume requirements.
CrystalChip and Counter-Diffusion Crystallization
The CrystalChip is a microfluidic platform that leverages the principle of counter-diffusion to create stable, convection-free concentration gradients ideal for high-quality crystal growth [82]. Its key advantage is dramatic sample reduction.
- Efficient Screening: With only 5 chips (requiring a total of 25 µL of purified protein), a researcher can screen 40 distinct precipitant gradients [82]. This efficiency is possible because each of the chip's 8 channels is loaded with a different concentrated crystallizing solution, which diffuses to create a continuum of conditions [82].
- In Situ Analysis: Crystals grown in the chip can be analyzed directly in situ using X-ray crystallography or optical methods, eliminating crystal handling damage and preserving diffraction quality [82]. The chip is compatible with synchrotron beamlines for direct data collection.
Impact on Serial Crystallography
The move towards miniaturization is especially critical for serial crystallography (SX) at synchrotrons and X-ray free-electron lasers (XFELs), a technique notorious for high sample consumption. Recent reviews highlight that advances in fixed-target and liquid injection microfluidic methods have reduced sample consumption from gram quantities in early SX experiments down to the microgram range [83]. The theoretical minimum for a full SX dataset is estimated to be as low as 450 ng of protein, a target that modern microfluidic delivery systems are steadily approaching [83].
Integrated Experimental Protocols
The true power of these technologies is realized when they are combined into a seamless, integrated workflow. The following protocols demonstrate how to implement AI-driven analysis and microfluidic screening in a complementary manner.
Protocol 4.1: AI-Assisted High-Throughput Screening in 96-Well Plates
This protocol uses sitting drop vapor diffusion in a 96-well format, coupled with automated imaging and AI scoring [80] [84].
Materials & Reagents
- Protein sample(s) of interest
- 96-well crystallization plate (e.g., SWISSCI 3 Lens Crystallisation Plate)
- Pre-packaged 96-condition crystallization screen (e.g., Morpheus HT-96)
- VIAFLO 96/384 handheld electronic pipette with a 96-channel head (50 µL) and low-retention GRIPTIPS [84]
- Sealing film or tape
- Rock Imager or compatible automated imaging system
- Rock Maker software with MARCO AI scoring license [80]
Procedure
- Transfer of Screening Agents:
- Load the VIAFLO 96 with tips. Aspirate 40 µL of the crystallization screen reagents from the 96-deep well block.
- Dispense 2 µL back into the source block (optional pre-dispense), then dispense 2 µL into each of the three lens wells and 30 µL into the reservoir well of the crystallization plate [84]. Discard tips.
Transfer of Protein Solutions:
- For up to three different protein solutions, load 8 tips at a time on the pipetting head. Aspirate the first protein solution from PCR strips placed in a cooling block.
- Dispense 2 µL of protein solution into the first lens well of each column. Repeat the process with the remaining protein solutions for the second and third lens wells, using new tips each time [84].
Sealing and Incubation:
- Seal the crystallization plate thoroughly to ensure proper vapor diffusion equilibrium.
- Incubate the plate at the desired temperature.
Automated Imaging and AI Scoring:
- Place the plate in the Rock Imager and schedule regular imaging inspections.
- In Rock Maker, initiate the MARCO auto-scoring function. The software will process the images, classifying each drop outcome in minutes [80].
- Manually review and validate the AI-generated scores, focusing on high-probability "Crystal" hits.
Protocol 4.2: Microfluidic Screening with CrystalChip
This protocol uses the counter-diffusion technique in a microfluidic format for efficient screening and sample conservation [82].
Materials & Reagents
- CrystalChip Discovery Screening Kit (includes 5 chips, pipette tips, holder, and 40 crystallization solutions)
- Purified protein sample
- 10 µL pipette and compatible low-retention tips (provided in kit)
Procedure
- Chip and Solution Preparation:
- Select a CrystalChip from the kit. Load the 8 reservoir channels with the desired concentrated crystallizing solutions (0.5-1 µL per reservoir) [82].
Protein Loading:
- Pipette 5 µL of purified protein into the single injection port of the chip [82]. The protein will fill the channels via capillary action.
Incubation and Crystal Growth:
- Place the chip in its holder and store it undisturbed at a constant temperature. The counter-diffusion process will create dynamic gradients, promoting nucleation and crystal growth over hours to weeks.
In Situ Imaging and Analysis:
- Monitor crystal growth directly by placing the entire chip on a microscope stage or in a crystal imager.
- For X-ray data collection, mount the chip in its dedicated holder on a goniometer at the synchrotron beamline. Translate the chip to center individual crystals in the beam for in situ data collection [82].
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful implementation of advanced crystallization workflows relies on a suite of specialized tools and reagents.
Table 2: Key Research Reagent Solutions for High-Throughput Crystallization
| Item | Function/Application | Example Product/Brand |
|---|---|---|
| Crystallization Plates | Platform for sitting drop vapor diffusion experiments; often designed for optimal imaging. | SWISSCI 3 Lens Crystallisation Plate [84] |
| Pre-Packaged Screens | Comprehensive sets of pre-mixed crystallization conditions for initial screening. | Morpheus HT-96 [84] |
| Electronic Pipettes | High-precision, automated liquid handling for rapid and reproducible plate setup. | INTEGRA VIAFLO 96/384 [84] |
| Microfluidic Chips | Miniaturized platform for counter-diffusion crystallization, drastically reducing sample consumption. | CrystalChip [82] |
| AI Scoring Software | Machine learning-based analysis of crystallization images for high-throughput scoring. | Rock Maker with MARCO [80] |
| Automated Imagers | Systems for scheduled, hands-off imaging of crystallization trials over time. | Rock Imager [80] |
Workflow Integration and Data Management
The integration of AI and microfluidics creates a new, streamlined workflow that fundamentally changes the research and development timeline in structural biology. The following diagram visualizes this integrated pathway, highlighting the parallel tracks and decision points that enhance efficiency.
This integrated workflow demonstrates a data-driven cycle. Failed conditions from either path contribute to a centralized database. Over time, this repository becomes a valuable asset for training more sophisticated AI models, which can, in turn, suggest more successful initial conditions for novel proteins, creating a virtuous cycle of continuous improvement [81] [80].
The fusion of AI integration and microfluidic miniaturization marks a definitive turning point for high-throughput protein crystallization. These technologies are not standalone solutions but are deeply synergistic. AI brings speed and intelligence to experimental design and analysis, while microfluidics enables feasibility and sustainability by conserving precious samples. For the research and drug development community, this means accelerated timelines, reduced costs, and the ability to tackle structurally challenging proteins that were previously intractable. As these tools continue to evolve—with AI models becoming more predictive and microfluidic devices more integrated with downstream analysis—the vision of a fully automated, sample-efficient pipeline for determining atomic-level structures is rapidly becoming a standard practice in modern structural biology.
Conclusion
High-throughput protein crystallization has evolved from a manual, artisanal process into a sophisticated, automated, and data-driven science essential for modern drug discovery and structural biology. By integrating foundational principles with advanced automation, AI-driven optimization, and specialized techniques for difficult targets, researchers can systematically overcome the historic bottleneck of crystal production. The convergence of robotic systems, miniaturized technologies, and intelligent software is paving the way for even greater throughput and success rates. As these technologies mature and become more accessible, they promise to deepen our understanding of disease mechanisms and dramatically accelerate the development of novel therapeutics, solidifying the role of structural biology as a cornerstone of biomedical innovation.
References
- 1. Review High-throughput protein crystallization [https://www.sciencedirect.com/science/article/abs/pii/S0959440X00001317]
- 2. Lessons from high-throughput protein crystallization ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3637395/]
- 3. 20 years of crystal hits: progress and promise in ultrahigh ... [https://journals.iucr.org/paper?cb5142]
- 4. High-Throughput Crystallization [https://www.creative-biostructure.com/high-throughput-crystallization-90.htm?srsltid=AfmBOoo7f8pSFPhgqfWffvcmkZVdh7UepDL4VkMJJVKIeznWd5D2g6Po]
- 5. High Throughput Protein Crystallization [https://ruppweb.org/Xray/tutorial/High_Throughput_EMBL_full.htm]
- 6. High-Throughput Protein Crystallization [https://www.sciencedirect.com/science/article/abs/pii/S1876162309770014]
- 7. Protein crystallization [https://en.wikipedia.org/wiki/Protein_crystallization]
- 8. Guide to Automated Protein Crystallization [https://formulatrix.com/life-science-automation-blog/automated-protein-crystallization-formulatrix/]
- 9. Protein Crystallization Market Report 2025: Projected $2.8 [https://www.globenewswire.com/news-release/2025/06/24/3104062/28124/en/Protein-Crystallization-Market-Report-2025-Projected-2-8-Billion-Growth-by-2029.html]
- 10. What Is Driving Global Protein Crystallization Market Growth [https://www.openpr.com/news/4281472/what-is-driving-global-protein-crystallization-market-growth]
- 11. Protein Crystallization Market Report 2024-2029, Featuring ... [https://finance.yahoo.com/news/protein-crystallization-market-report-2024-081400725.html]
- 12. Protein Crystallization Market By Size, Share and Forecast ... [https://www.techsciresearch.com/report/protein-crystallization-market/23962.html]
- 13. Protein Crystallization Market Size & Global Trends |2033 [https://straitsresearch.com/report/protein-crystallization-market]
- 14. Protein Crystallization Market Size, Growth, Trends & ... [https://www.mordorintelligence.com/industry-reports/protein-crystallization-market]
- 15. Latest Forecast Shows Protein Crystallization and ... [https://www.linkedin.com/pulse/latest-forecast-shows-protein-crystallization-crystallography-qecfe]
- 16. Protein Crystallization & Crystallography Market 2025-2032 [https://www.360iresearch.com/library/intelligence/protein-crystallization-crystallography]
- 17. Protein Crystallization for X-ray Crystallography - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3182643/]
- 18. Mechanisms of membrane protein crystallization in 'bicelles' [https://www.nature.com/articles/s41598-022-13945-0]
- 19. advances and challenges in controlling protein nucleation [https://pubs.rsc.org/en/content/articlehtml/2025/ce/d5ce00767d]
- 20. From Solution to Crystal: Mastering Protein Crystallization [https://www.creative-biostructure.com/resource-protein-crystallization-guide.htm?srsltid=AfmBOorLuUyJwwtndDM_ErhwRTgtYjwRMFcXUMMfowQsCiRnRU4emxIY]
- 21. Protein Crystallization: Hanging & Sitting Drops [https://www.proteinstructures.com/protein-crystallization-basics/]
- 22. COMPARISON OF MICROBATCH AND VAPOR DIFFUSION [https://www.douglas.co.uk/rep3.htm]
- 23. The advantages of using a modified microbatch method for ... [https://pubmed.ncbi.nlm.nih.gov/12554964/]
- 24. Automated Protocols for Macromolecular Crystallization at ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5908693/]
- 25. Protein Crystallization Automation and Software [https://formulatrix.com/protein-crystallization-systems/]
- 26. Automated systems for protein crystallization [https://www.sciencedirect.com/science/article/abs/pii/S104620230400115X]
- 27. Automation of protein crystallization scaleup via Opentrons ... [https://www.sciencedirect.com/science/article/pii/S2472630325000263]
- 28. Automated Crystallization Imaging System [https://pharma-industry-review.com/automated-crystallization-imaging-system]
- 29. High-Throughput Protein Crystallography [https://www.sciencedirect.com/science/article/pii/S1535553505003138]
- 30. Automation in biological crystallization - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC4051518/]
- 31. mosquito crystal [https://www.sptlabtech.com/products/mosquito/mosquito-crystal]
- 32. TTP SPT Labtech Dragonfly Crystal Non-Contact Liquid ... [https://www.bostonind.com/ttp-spt-labtech-dragonfly-crystal-non-contact-liquid-handler-protein-crystallization,-10-dispensing-heads?srsltid=AfmBOoq6Y2-FiK3yIceZ3t8715k-3RpW74_lZrSauNEeN7Dd_q2bih6E]
- 33. Rock Imager - Protein Crystallization [https://formulatrix.com/protein-crystallization-systems/rock-imager-protein-images/]
- 34. Automated storage and visual inspection of crystal plates ... [https://bic.ceitec.cz/automated-storage-and-visual-inspection-of-crystal-plates-4-c-and-20-c/]
- 35. Protein Crystallization Screening - Automation Startup ... [https://formulatrix.com/protein-crystallization-systems/protein-crystallization-screening-automation-bundle/]
- 36. Pac@Van: Protein Automated Crystallography @ Vanderbilt [https://www.vanderbilt.edu/csb/facilities/labs-instrumentation-facility/biomolecular-crystallography/pacvan-protein-automated-crystallography-vanderbilt/]
- 37. Some practical guidelines for UV imaging in the protein ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3564629/]
- 38. Multi-Fluorescence Imaging - for Protein Crystallization [https://formulatrix.com/multi-fluorescence-imaging/]
- 39. SONICC 纳微米晶体检测系统 [https://formulatrix.tech/protein-crystallization-systems/sonicc-%E7%BA%B3%E5%BE%AE%E7%B1%B3%E6%99%B6%E4%BD%93%E6%A3%80%E6%B5%8B%E7%B3%BB%E7%BB%9F/]
- 40. Precision Imaging for Protein Crystallization: Real-World ... [https://formulatrix.com/life-science-automation-blog/precision-imaging-for-protein-crystallization/]
- 41. Crystals | Special Issue : Membrane Protein Crystallography [https://www.mdpi.com/journal/crystals/special_issues/membraneprotein]
- 42. Membrane Protein Crystallisation: Current Trends and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5033070/]
- 43. High-Throughput Crystallization platform - [https://www.itaca-sb.it/high-throughput-crystallization-platform/]
- 44. Computational design of soluble and functional membrane ... [https://www.nature.com/articles/s41586-024-07601-y]
- 45. Optimization of crystallization conditions for biological ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4231845/]
- 46. Iterative screen optimization maximizes the efficiency of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6360444/]
- 47. Preparing for successful protein crystallization experiments [https://pmc.ncbi.nlm.nih.gov/articles/PMC12210191/]
- 48. Enhancing Protein Crystallization through Precipitant ... [https://www.sciencedirect.com/science/article/pii/S0969212603001850]
- 49. High Throughput pH Optimization of Protein Crystallization [https://link.springer.com/protocol/10.1007/978-1-60327-058-8_27]
- 50. High‐Throughput Pipeline for Protein Expression and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12306927/]
- 51. Cooling-rate screening system for determining protein ... [https://www.sciencedirect.com/science/article/abs/pii/S0022024806003502]
- 52. High-Throughput Screening to Obtain Crystal Hits for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10259656/]
- 53. Seeds to crystals [https://www.sciencedirect.com/science/article/abs/pii/S104784770300039X]
- 54. 5 Protein Crystallization Seeding Methods for Bigger Crystals [https://bitesizebio.com/59302/protein-crystallization-seeding/]
- 55. Seeding - Terese Bergfors [https://xray.teresebergfors.com/seeding/]
- 56. 'Seeding' with protease to optimize protein crystallization ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3374523/]
- 57. Improve co-crystal screening using machine learning and ... [https://www.crystallizationsystems.com/news/improve-co-crystal-screening-using-machine-learning-and-crystal16/]
- 58. AI Learns to Uncover the Hidden Atomic Structure of Crystals [https://www.engineering.columbia.edu/about/news/ai-learns-uncover-hidden-atomic-structure-crystals]
- 59. AI model can reveal the structures of crystalline materials [https://science.mit.edu/ai-model-can-reveal-the-structures-of-crystalline-materials/]
- 60. High-throughput crystallisation and ligand screening [https://www.embl.org/services-facilities/grenoble/high-throughput-crystallisation/]
- 61. Salvage of Failed Protein Targets by Reductive Alkylation [https://pmc.ncbi.nlm.nih.gov/articles/PMC4078742/]
- 62. Genome Pool Strategy for Structural Coverage of Protein ... [https://www.sciencedirect.com/science/article/pii/S0969212608003742]
- 63. Emerging microfluidic platforms for crystallization process ... [https://www.sciencedirect.com/science/article/abs/pii/S0263876223005191]
- 64. Small scale, big results: The advantages of microfabricated ... [https://www.ufluidix.com/blog/small-scale-big-results-the-advantages-of-microfabricated-devices-for-protein-crystallization/]
- 65. A digital microfluidic approach to increasing sample ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11849296/]
- 66. National Crystallization Center [https://www.buffalo.edu/hwi/research/national-crystallization-center.html]
- 67. Benchmarking protein language models for ... [https://www.nature.com/articles/s41598-025-86519-5]
- 68. Survey of predictors of propensity for protein production ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7001581/]
- 69. Protein crystallization screens developed at the MRC ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4911435/]
- 70. Efficiency analysis of sampling protocols used in protein ... [https://www.sciencedirect.com/science/article/abs/pii/S002202480101154X]
- 71. Protein XRD Protocols - Crystallization of Proteins [https://sites.google.com/colgate.edu/xrd-protocols/home/crystallization-of-proteins]
- 72. High-resolution protein modeling through Cryo-EM and AI [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590954/]
- 73. High-throughput protein crystallography to empower ... [https://pubmed.ncbi.nlm.nih.gov/40237633/]
- 74. Automated model building and protein identification in cryo ... [https://www.nature.com/articles/s41586-024-07215-4]
- 75. High-Throughput Screening Core | Research Resources Center [https://rrc.uic.edu/cores/bioanalytics-biophysics-cytomics/high-throughput-screening-core/]
- 76. Research team sheds light on the future potential of X-ray ... [https://www.controlled-molecule-imaging.org/news/news/2025/research_team_sheds_light_on_the_future_potential_of_x_ray_imaging_and_cryo_electron_microscopy/]
- 77. Optimizing Associative Experimental for Design ... Protein [https://pubmed.ncbi.nlm.nih.gov/26955046/]
- 78. (PDF) Protein Screening Using Associative... Crystallization [https://www.academia.edu/33267300/Protein_Crystallization_Screening_Using_Associative_Experimental_Design]
- 79. Protein Crystallography Market Overview 2025: Key Trends ... [https://www.linkedin.com/pulse/protein-crystallography-market-overview-2025-key-trends-nbvce/]
- 80. Protein Crystallization Software - ROCK MAKER® [https://formulatrix.com/protein-crystallization-systems/rock-maker-protein-crystallization-software/]
- 81. Protein Crystallization Automation Workshop 2025 - FORMULATRIX [https://formulatrix.com/life-science-automation-blog/protein-crystallization-automation-workshop-2025/]
- 82. CrystalChip Microfluidic Chips For Protein Crystallization [https://www.mitegen.com/product/crystalchip-microfluidic-chips-for-protein-crystallization/]
- 83. Sample delivery methods for protein X-ray crystallography ... [https://www.nature.com/articles/s41467-025-65173-5]
- 84. Set-up of protein crystallization plates with the VIAFLO 96 [https://www.integra-biosciences.com/united-states/en/applications/easy-set-high-throughput-protein-crystallization-experiments-using-viaflo-96-and-viaflo-384-handheld-electronic-pipette]